Abstract
The title compound, C33H35N3O5, is the product of the multicomponent condensation of 1-benzyl-4-ethoxycarbonylpiperidin-3-one with 1,5-bis(2-formylphenoxy)-3-oxapentane and ammonium acetate. The molecule comprises a pentacyclic system containing the aza-14-crown-4-ether macrocycle, tetrahydropyrimidine, tetrahydropyridine and two benzene rings. The aza-14-crown-4-ether ring adopts a bowl conformation with a dihedral angle of 62.37 (5)° between the benzene rings. The tetrahydropyrimidine ring has an envelope conformation with the chiral C atom as the flap, whereas the tetrahydropyridine ring adopts a distorted chair conformation. Two amino groups are involved in intramolecular N—H⋯O hydrogen bonds. In the crystal, weak C—H⋯O hydrogen bonds link the molecules into layers parallel to the ab plane.
Related literature
For general background to the design, synthesis, chemical properties and applications of macrocyclic ligands for coordination chemistry, see: Hiraoka (1982 ▶); Pedersen (1988 ▶); Gokel & Murillo (1996 ▶); Bradshaw & Izatt (1997 ▶). For the crystal structures of related compounds, see: Levov et al. (2006 ▶, 2008 ▶); Komarova et al. (2008 ▶); Anh et al. (2008 ▶, 2012a
▶,b
▶,c
▶); Hieu et al. (2009 ▶, 2011 ▶, 2012a
▶,b
▶); Khieu et al. (2011 ▶); Sokol et al. (2011 ▶).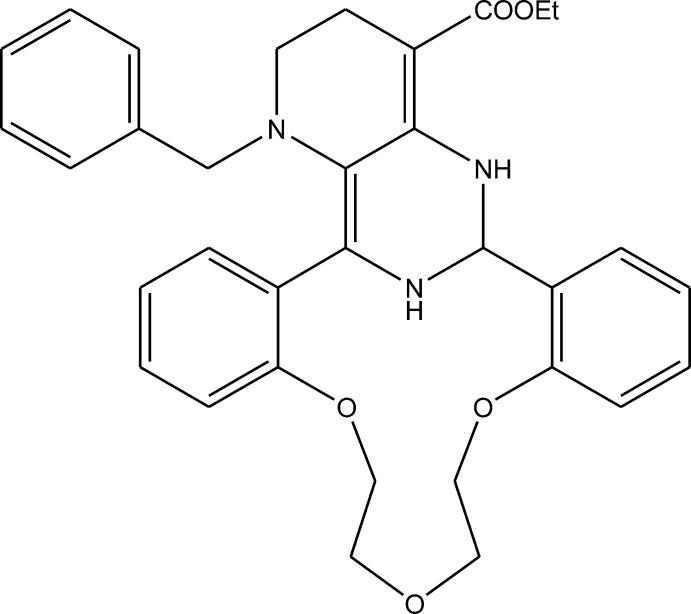
Experimental
Crystal data
C33H35N3O5
M r = 553.64
Monoclinic,

a = 10.5304 (5) Å
b = 12.6363 (5) Å
c = 10.7246 (5) Å
β = 92.865 (1)°
V = 1425.29 (11) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 100 K
0.30 × 0.24 × 0.21 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2003 ▶) T min = 0.974, T max = 0.982
18837 measured reflections
8289 independent reflections
6878 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.094
S = 1.00
8289 reflections
377 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.23 e Å−3
Δρmin = −0.18 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813007241/cv5393sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813007241/cv5393Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813007241/cv5393Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N24—H24⋯O1′ | 0.882 (18) | 2.015 (18) | 2.6928 (16) | 132.8 (15) |
| N25—H25⋯O14 | 0.882 (18) | 2.441 (17) | 2.9744 (17) | 119.3 (13) |
| C6—H6⋯O1′i | 0.95 | 2.42 | 3.3516 (19) | 168 |
| C18—H18⋯O1′ii | 0.95 | 2.42 | 3.3613 (19) | 174 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
We thank the National Foundation for Science and Technology Development (NAFOSTED), Hanoi, Vietnam (grant No. 104.02–2012.44) for the financial support of this work.
supplementary crystallographic information
Comment
Design, preparation and applications of macroheterocyclic ligands for coordination, supramolecular and medicinal chemistry draw constant attention of investigators during the last several decades (Hiraoka, 1982; Pedersen,1988; Gokel & Murillo, 1996; Bradshaw & Izatt, 1997). Recently we have developed the effective method of synthesis of azacrown ethers including piperidine (Levov et al., 2006); Anh et al., 2008, 2012a,b), cycloalkanopiperidine (Levov et al. 2008); bispidine (Komarova et al.; Sokol et al.; Hieu et al. 2012a; Anh et al. 2012c); perhydropyrimidine (Hieu et al., 2011) and perhydrotriazine (Hieu et al., 2009, 2012b; Khieu et al., 2011) subunits.
In attempts to apply the chemistry for obtaining azacrown ether containing ethoxy-substituted bispidino subunit with two nitrogen atoms in the unsymmetrical positions, we studied the multicomponent condensation of the 1-benzyl-4-ethoxycarbonylpiperidin-3-one (ketone component) with 1,5-bis(2-formylphenoxy)-3-oxapentane (podand) and ammonium acetate. The reaction has proceeded smoothly under mild conditions to give the title compound as an unexpected product (Fig. 1). The first step of this cascade process appears to be the intermolecular condensation of one aldehyde group of the podand with the activated methylene group of the ketone component. Then the addition of one molecule of ammonia to the keto-group yields its hydroxyl-amino form. Further the second aldehyde group is condensed with the amino group to form the intermediate azacrown ether containing 1,4-azadiene fragment fused to piperidine moiety. The final step is the double Mannich cycloaddition of another molecule of ammonia to the azadiene moiety followed by dehydration to form the product. The structure of the new azacrown system, C33H35N3O5 (I), was unambiguously established by X-ray diffraction study.
The molecule of I comprises a pentacyclic system containing the aza-14-crown-4-ether macrocycle, tetrahydropyrimidine, tetrahydropyridine and two benzene rings (Fig. 2). The aza-14-crown-4-ether ring adopts a bowl conformation. The configuration of the C7—O8—C9—C10 —O11—C12—C13—O14—C15 polyether chain is t–g(-)–t–t–g(+)–t (t = trans, 180°; g = gauche, ±60°). The dihedral angle between the planes of the benzene rings fused to the aza-14-crown-4-ether moiety is 62.37 (5)°. The central tetrahydropyrimidine ring has an envelope conformation (the C1 carbon atom is out of the plane passed through the other atoms of the ring (r.m.s. deviation = 0.023 Å) by 0.661 (2) Å), which is stabilized by the intramolecular N25—H25···O14 hydrogen bond (Table 1). The terminal tetrahydropyridine ring adopts a distorted chair conformation (the N1' nitrogen and C6' carbon atoms are out of the plane passed through the other atoms of the ring (r.m.s. deviation = 0.012 Å) by -0.245 (3) and 0.431 (3) Å, respectively). The three N24, N25 and N1' nitrogen atoms have the trigonal-pyramidal geometries. The carboxylate substituent (except for the terminal C16' carbon atom) is practically coplanar to the basal C22—C23—C4'—C5' plane of the tetrahydropyridine ring (the O2'—C14'—C4'—C5' dihedral angle is -5.5 (2)°). This disposition is apparently determined by the intramolecular N24—H24···O1' hydrogen bond (Table 1).
The molecule of I possesses an asymmetric center at the C1 carbon atom and crystallizes in the chiral space group P21. However, its absolute configuration cannot be objectively determined because the absence of the heavy (Z > 14) atoms within the molecule.
In the crystal, the molecules of I are bound by the weak intermolecular C—H···O hydrogen bonding interactions (Table 1) into layers parallel to ab> plane (Figure 3).
Experimental
Ammonium acetate (5.0 g, 65 mmol) was added to a solution of 1,5-bis(2-formylphenoxy)-3-oxapentane (1.57 g, 5.0 mmol) and 1-benzyl-4-ethoxycarbonylpiperidin-3-one (1.48 g, 5.0 mmol) in ethanol (30 ml) – acetic acid (2 ml). The reaction mixture was stirred at 293 K for 3 days. At the end of the reaction, the formed precipitate was filtered off, washed with ethanol and chromatographically purified on the column filled with silica gel. A re-crystallization from hexane:ethylacetate (3:1) mixture gave 0.83 g of light-yellow crystals of I.Yield is 30.0%. M.p. = 373–376 K. IR (KBr), ν/cm-1: 1599, 1644, 3297, 3374, 3453. 1H NMR (CDCl3, 400 MHz, 300 K): δ = 1.29 (t, 3H, J = 7.2 and 6.8, CH2CH3), 2.26 and 2.78 (both m, 1H and 3H, correspondingly, NCH2CH2), 3.50 and 3.85 (both d, 1H each, J = 13.2 each, NCH2Ar), 3.73–4.15 (m, 9H, OCH2CH2OCH2CH2O and CH2CH3), 4.83 (s, 1H, N—H25), 6.05 (s, 1H, H1), 6.73 (dd, 2H, J = 7.7 and 1.6, H6 and H16), 6.8 (broad t, 2H, J = 8.9, H4 and H18), 6.97–7.09 (m, 5H, Harom), 7.28–7.32 (m, 2H, Harom), 7.47 (dd, 2H, J = 7.6 and 1.6, H3), 7.87 (dd, 2H, J = 7.6 and 1.2, H19), 8.61 (s, 1H, N—H24). Anal. Calcd for C33H35N3O5: C, 71.59; H, 6.37; N, 7.59. Found: C, 71.53; H, 6.22; N, 7.37.
Refinement
The absolute structure of I cannot be objectively determined by the refinement of Flack parameter because the absence of the heavy (Z > 14) atoms within the molecule.
The hydrogen atoms of the amino groups were localized in the difference-Fourier map and refined isotropically with fixed isotropic displacement parameters [Uiso(H) = 1.2Ueq(N)]. The other hydrogen atoms were placed in calculated positions with C—H = 0.95–1.00 Å and refined in the riding model with fixed isotropic displacement parameters [Uiso(H) = 1.5Ueq(C) for the methyl group and 1.2Ueq(C) for the other groups].
Figures
Fig. 1.

Multicomponent condensation of the 1-benzyl-4-ethoxycarbonylpiperidin-3-one with 1,5-bis(2-formylphenoxy)-3-oxapentane and ammonium acetate.
Fig. 2.
Molecular structure of I. Displacement ellipsoids are shown at the 50% probability level. H atoms are presented as small spheres of arbitrary radius. The intramolecular N—H···O hydrogen bonds are drawn by dashed lines.
Fig. 3.
A portion of the crystal structure showing the weak intermolecular C—H···O hydrogen bonds, which are depicted by dashed lines.
Crystal data
| C33H35N3O5 | F(000) = 588 |
| Mr = 553.64 | Dx = 1.290 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 5715 reflections |
| a = 10.5304 (5) Å | θ = 2.5–31.8° |
| b = 12.6363 (5) Å | µ = 0.09 mm−1 |
| c = 10.7246 (5) Å | T = 100 K |
| β = 92.865 (1)° | Prism, yellow |
| V = 1425.29 (11) Å3 | 0.30 × 0.24 × 0.21 mm |
| Z = 2 |
Data collection
| Bruker APEXII CCD diffractometer | 8289 independent reflections |
| Radiation source: fine-focus sealed tube | 6878 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.0°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2003) | h = −14→14 |
| Tmin = 0.974, Tmax = 0.982 | k = −17→17 |
| 18837 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.094 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0431P)2 + 0.123P] where P = (Fo2 + 2Fc2)/3 |
| 8289 reflections | (Δ/σ)max < 0.001 |
| 377 parameters | Δρmax = 0.23 e Å−3 |
| 1 restraint | Δρmin = −0.18 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.22606 (12) | 0.22945 (12) | 0.85547 (12) | 0.0266 (3) | |
| H1 | 0.1992 | 0.2950 | 0.8090 | 0.032* | |
| C2 | 0.16248 (12) | 0.22544 (12) | 0.97887 (12) | 0.0271 (3) | |
| C3 | 0.13663 (14) | 0.13054 (13) | 1.03713 (13) | 0.0310 (3) | |
| H3 | 0.1570 | 0.0660 | 0.9974 | 0.037* | |
| C4 | 0.08137 (14) | 0.12737 (14) | 1.15267 (14) | 0.0347 (3) | |
| H4 | 0.0644 | 0.0615 | 1.1912 | 0.042* | |
| C5 | 0.05172 (14) | 0.22147 (15) | 1.21021 (14) | 0.0376 (3) | |
| H5 | 0.0148 | 0.2200 | 1.2892 | 0.045* | |
| C6 | 0.07508 (15) | 0.31771 (14) | 1.15432 (14) | 0.0365 (3) | |
| H6 | 0.0533 | 0.3819 | 1.1942 | 0.044* | |
| C7 | 0.13097 (13) | 0.31993 (13) | 1.03861 (13) | 0.0312 (3) | |
| O8 | 0.15789 (13) | 0.41119 (10) | 0.97691 (11) | 0.0441 (3) | |
| C9 | 0.16598 (16) | 0.50775 (12) | 1.04698 (14) | 0.0360 (3) | |
| H9A | 0.2106 | 0.4957 | 1.1292 | 0.043* | |
| H9B | 0.0799 | 0.5356 | 1.0607 | 0.043* | |
| C10 | 0.23872 (16) | 0.58408 (13) | 0.97119 (14) | 0.0367 (3) | |
| H10A | 0.2017 | 0.5874 | 0.8846 | 0.044* | |
| H10B | 0.2362 | 0.6558 | 1.0081 | 0.044* | |
| O11 | 0.36498 (10) | 0.54650 (9) | 0.97231 (10) | 0.0380 (3) | |
| C12 | 0.43821 (16) | 0.59053 (13) | 0.87902 (14) | 0.0372 (3) | |
| H12A | 0.4594 | 0.6651 | 0.8995 | 0.045* | |
| H12B | 0.3899 | 0.5889 | 0.7975 | 0.045* | |
| C13 | 0.55719 (15) | 0.52654 (12) | 0.87280 (14) | 0.0338 (3) | |
| H13A | 0.6134 | 0.5571 | 0.8108 | 0.041* | |
| H13B | 0.6039 | 0.5252 | 0.9552 | 0.041* | |
| O14 | 0.51863 (10) | 0.42183 (9) | 0.83650 (11) | 0.0372 (2) | |
| C15 | 0.61093 (14) | 0.35024 (12) | 0.81031 (13) | 0.0298 (3) | |
| C16 | 0.74063 (14) | 0.37340 (13) | 0.81731 (15) | 0.0361 (3) | |
| H16 | 0.7694 | 0.4404 | 0.8472 | 0.043* | |
| C17 | 0.82748 (14) | 0.29860 (14) | 0.78062 (15) | 0.0388 (4) | |
| H17 | 0.9156 | 0.3150 | 0.7847 | 0.047* | |
| C18 | 0.78697 (14) | 0.20051 (13) | 0.73815 (15) | 0.0367 (4) | |
| H18 | 0.8465 | 0.1499 | 0.7114 | 0.044* | |
| C19 | 0.65764 (14) | 0.17656 (13) | 0.73505 (14) | 0.0325 (3) | |
| H19 | 0.6300 | 0.1085 | 0.7075 | 0.039* | |
| C20 | 0.56790 (13) | 0.25002 (12) | 0.77133 (12) | 0.0273 (3) | |
| C21 | 0.43077 (12) | 0.22008 (12) | 0.76601 (12) | 0.0269 (3) | |
| C22 | 0.37341 (13) | 0.17558 (12) | 0.66288 (13) | 0.0272 (3) | |
| C23 | 0.24979 (13) | 0.12495 (11) | 0.66886 (12) | 0.0255 (3) | |
| N24 | 0.19281 (12) | 0.13681 (10) | 0.78031 (11) | 0.0289 (3) | |
| H24 | 0.1137 (17) | 0.1140 (14) | 0.7817 (16) | 0.035* | |
| N25 | 0.36503 (11) | 0.22816 (11) | 0.87544 (11) | 0.0278 (2) | |
| H25 | 0.3951 (16) | 0.2792 (14) | 0.9247 (16) | 0.033* | |
| N1' | 0.43614 (11) | 0.17458 (10) | 0.54726 (10) | 0.0272 (2) | |
| C4' | 0.20023 (13) | 0.06475 (12) | 0.57082 (12) | 0.0280 (3) | |
| C5' | 0.27720 (14) | 0.04519 (14) | 0.45751 (13) | 0.0353 (3) | |
| H5A | 0.2425 | 0.0885 | 0.3869 | 0.042* | |
| H5B | 0.2699 | −0.0302 | 0.4331 | 0.042* | |
| C6' | 0.41709 (14) | 0.07292 (12) | 0.48401 (13) | 0.0306 (3) | |
| H6A | 0.4575 | 0.0165 | 0.5364 | 0.037* | |
| H6B | 0.4602 | 0.0747 | 0.4041 | 0.037* | |
| C7' | 0.39864 (15) | 0.26627 (13) | 0.46972 (15) | 0.0357 (3) | |
| H7A | 0.3881 | 0.3282 | 0.5248 | 0.043* | |
| H7B | 0.3151 | 0.2514 | 0.4269 | 0.043* | |
| C8' | 0.49217 (14) | 0.29427 (11) | 0.37288 (13) | 0.0301 (3) | |
| C9' | 0.44865 (17) | 0.34402 (13) | 0.26331 (15) | 0.0399 (4) | |
| H9 | 0.3599 | 0.3525 | 0.2462 | 0.048* | |
| C10' | 0.5333 (2) | 0.38131 (15) | 0.17872 (17) | 0.0510 (5) | |
| H10 | 0.5024 | 0.4164 | 0.1049 | 0.061* | |
| C11' | 0.6611 (2) | 0.36766 (15) | 0.20134 (18) | 0.0542 (5) | |
| H11 | 0.7189 | 0.3930 | 0.1431 | 0.065* | |
| C12' | 0.70637 (18) | 0.31703 (15) | 0.30879 (18) | 0.0474 (4) | |
| H12 | 0.7952 | 0.3070 | 0.3239 | 0.057* | |
| C13' | 0.62174 (15) | 0.28063 (13) | 0.39512 (15) | 0.0366 (3) | |
| H13 | 0.6531 | 0.2464 | 0.4693 | 0.044* | |
| C14' | 0.07571 (14) | 0.01805 (12) | 0.57658 (13) | 0.0298 (3) | |
| O1' | 0.00688 (10) | 0.02090 (9) | 0.66562 (9) | 0.0342 (2) | |
| O2' | 0.03869 (11) | −0.03237 (11) | 0.46902 (11) | 0.0452 (3) | |
| C15' | −0.08813 (17) | −0.07689 (17) | 0.46202 (18) | 0.0507 (5) | |
| H15A | −0.1111 | −0.1000 | 0.5462 | 0.061* | |
| H15B | −0.0901 | −0.1397 | 0.4068 | 0.061* | |
| C16' | −0.1818 (2) | 0.0022 (2) | 0.4129 (3) | 0.0858 (9) | |
| H16A | −0.2654 | −0.0313 | 0.4006 | 0.129* | |
| H16B | −0.1548 | 0.0298 | 0.3330 | 0.129* | |
| H16C | −0.1871 | 0.0605 | 0.4727 | 0.129* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0254 (6) | 0.0334 (7) | 0.0211 (6) | 0.0014 (6) | 0.0027 (5) | −0.0038 (5) |
| C2 | 0.0213 (6) | 0.0390 (7) | 0.0210 (6) | 0.0017 (6) | 0.0012 (5) | −0.0044 (6) |
| C3 | 0.0274 (7) | 0.0388 (8) | 0.0271 (7) | 0.0060 (6) | 0.0032 (5) | −0.0015 (6) |
| C4 | 0.0281 (7) | 0.0466 (9) | 0.0299 (7) | 0.0044 (6) | 0.0053 (6) | 0.0039 (6) |
| C5 | 0.0307 (7) | 0.0566 (10) | 0.0262 (7) | 0.0022 (7) | 0.0087 (5) | −0.0027 (7) |
| C6 | 0.0354 (8) | 0.0456 (9) | 0.0295 (7) | 0.0015 (7) | 0.0089 (6) | −0.0103 (7) |
| C7 | 0.0298 (7) | 0.0385 (8) | 0.0257 (7) | −0.0007 (6) | 0.0045 (5) | −0.0063 (6) |
| O8 | 0.0656 (8) | 0.0368 (6) | 0.0311 (6) | −0.0050 (6) | 0.0139 (5) | −0.0102 (5) |
| C9 | 0.0405 (8) | 0.0359 (8) | 0.0317 (7) | 0.0062 (6) | 0.0044 (6) | −0.0091 (6) |
| C10 | 0.0431 (9) | 0.0337 (8) | 0.0329 (8) | 0.0110 (7) | −0.0015 (6) | −0.0025 (6) |
| O11 | 0.0395 (6) | 0.0419 (6) | 0.0330 (5) | 0.0087 (5) | 0.0049 (5) | 0.0092 (5) |
| C12 | 0.0484 (9) | 0.0305 (7) | 0.0327 (8) | 0.0005 (7) | 0.0014 (7) | 0.0052 (6) |
| C13 | 0.0386 (8) | 0.0323 (8) | 0.0306 (7) | −0.0076 (6) | 0.0026 (6) | 0.0011 (6) |
| O14 | 0.0305 (5) | 0.0331 (6) | 0.0484 (7) | −0.0034 (4) | 0.0060 (5) | −0.0088 (5) |
| C15 | 0.0286 (7) | 0.0345 (7) | 0.0267 (7) | −0.0018 (6) | 0.0049 (5) | 0.0024 (5) |
| C16 | 0.0315 (8) | 0.0398 (9) | 0.0370 (8) | −0.0073 (6) | 0.0027 (6) | 0.0032 (7) |
| C17 | 0.0251 (7) | 0.0517 (10) | 0.0399 (8) | −0.0032 (7) | 0.0043 (6) | 0.0146 (7) |
| C18 | 0.0283 (7) | 0.0469 (10) | 0.0353 (8) | 0.0096 (6) | 0.0059 (6) | 0.0089 (7) |
| C19 | 0.0322 (7) | 0.0368 (8) | 0.0288 (7) | 0.0029 (6) | 0.0035 (6) | 0.0033 (6) |
| C20 | 0.0252 (6) | 0.0358 (8) | 0.0211 (6) | −0.0013 (5) | 0.0034 (5) | 0.0027 (5) |
| C21 | 0.0244 (6) | 0.0309 (7) | 0.0257 (6) | −0.0001 (6) | 0.0046 (5) | −0.0027 (6) |
| C22 | 0.0266 (7) | 0.0336 (7) | 0.0220 (6) | −0.0012 (6) | 0.0070 (5) | −0.0004 (5) |
| C23 | 0.0260 (6) | 0.0287 (7) | 0.0220 (6) | 0.0024 (5) | 0.0026 (5) | −0.0005 (5) |
| N24 | 0.0261 (6) | 0.0375 (7) | 0.0237 (5) | −0.0049 (5) | 0.0061 (5) | −0.0066 (5) |
| N25 | 0.0242 (5) | 0.0367 (6) | 0.0229 (5) | −0.0029 (5) | 0.0039 (4) | −0.0067 (5) |
| N1' | 0.0292 (6) | 0.0331 (6) | 0.0197 (5) | 0.0018 (5) | 0.0057 (4) | 0.0014 (4) |
| C4' | 0.0285 (7) | 0.0341 (8) | 0.0216 (6) | 0.0000 (6) | 0.0020 (5) | −0.0031 (5) |
| C5' | 0.0343 (8) | 0.0487 (9) | 0.0230 (6) | −0.0027 (7) | 0.0043 (6) | −0.0086 (6) |
| C6' | 0.0305 (7) | 0.0380 (8) | 0.0236 (6) | 0.0027 (6) | 0.0058 (5) | −0.0037 (6) |
| C7' | 0.0349 (8) | 0.0405 (8) | 0.0323 (7) | 0.0109 (6) | 0.0092 (6) | 0.0078 (6) |
| C8' | 0.0372 (8) | 0.0272 (7) | 0.0265 (7) | 0.0017 (6) | 0.0064 (6) | 0.0013 (5) |
| C9' | 0.0499 (9) | 0.0385 (9) | 0.0311 (7) | 0.0039 (7) | 0.0001 (7) | 0.0061 (6) |
| C10' | 0.0774 (14) | 0.0410 (10) | 0.0352 (9) | −0.0028 (9) | 0.0092 (9) | 0.0128 (7) |
| C11' | 0.0757 (14) | 0.0426 (10) | 0.0466 (10) | −0.0157 (9) | 0.0262 (10) | 0.0039 (8) |
| C12' | 0.0409 (9) | 0.0468 (10) | 0.0555 (10) | −0.0095 (8) | 0.0138 (8) | −0.0014 (9) |
| C13' | 0.0377 (8) | 0.0378 (8) | 0.0346 (8) | 0.0006 (6) | 0.0047 (6) | 0.0034 (6) |
| C14' | 0.0307 (7) | 0.0312 (7) | 0.0273 (7) | −0.0001 (6) | −0.0015 (5) | −0.0019 (6) |
| O1' | 0.0327 (5) | 0.0388 (6) | 0.0314 (5) | −0.0058 (4) | 0.0052 (4) | −0.0018 (4) |
| O2' | 0.0381 (6) | 0.0640 (8) | 0.0335 (6) | −0.0157 (6) | 0.0017 (5) | −0.0158 (5) |
| C15' | 0.0457 (10) | 0.0629 (12) | 0.0430 (10) | −0.0233 (9) | −0.0026 (8) | −0.0109 (9) |
| C16' | 0.0484 (12) | 0.113 (2) | 0.0931 (19) | −0.0321 (13) | −0.0237 (12) | 0.0484 (17) |
Geometric parameters (Å, º)
| C1—N24 | 1.4542 (18) | C21—C22 | 1.3556 (19) |
| C1—N25 | 1.4685 (17) | C21—N25 | 1.3957 (16) |
| C1—C2 | 1.5135 (17) | C22—N1' | 1.4338 (17) |
| C1—H1 | 1.0000 | C22—C23 | 1.4548 (19) |
| C2—C3 | 1.386 (2) | C23—N24 | 1.3721 (17) |
| C2—C7 | 1.403 (2) | C23—C4' | 1.3790 (19) |
| C3—C4 | 1.396 (2) | N24—H24 | 0.882 (18) |
| C3—H3 | 0.9500 | N25—H25 | 0.883 (18) |
| C4—C5 | 1.383 (2) | N1'—C6' | 1.4620 (19) |
| C4—H4 | 0.9500 | N1'—C7' | 1.4687 (19) |
| C5—C6 | 1.383 (3) | C4'—C14' | 1.442 (2) |
| C5—H5 | 0.9500 | C4'—C5' | 1.5144 (19) |
| C6—C7 | 1.400 (2) | C5'—C6' | 1.527 (2) |
| C6—H6 | 0.9500 | C5'—H5A | 0.9900 |
| C7—O8 | 1.366 (2) | C5'—H5B | 0.9900 |
| O8—C9 | 1.4334 (18) | C6'—H6A | 0.9900 |
| C9—C10 | 1.497 (2) | C6'—H6B | 0.9900 |
| C9—H9A | 0.9900 | C7'—C8' | 1.509 (2) |
| C9—H9B | 0.9900 | C7'—H7A | 0.9900 |
| C10—O11 | 1.4114 (19) | C7'—H7B | 0.9900 |
| C10—H10A | 0.9900 | C8'—C13' | 1.384 (2) |
| C10—H10B | 0.9900 | C8'—C9' | 1.390 (2) |
| O11—C12 | 1.4079 (18) | C9'—C10' | 1.386 (2) |
| C12—C13 | 1.495 (2) | C9'—H9 | 0.9500 |
| C12—H12A | 0.9900 | C10'—C11' | 1.367 (3) |
| C12—H12B | 0.9900 | C10'—H10 | 0.9500 |
| C13—O14 | 1.4322 (18) | C11'—C12' | 1.382 (3) |
| C13—H13A | 0.9900 | C11'—H11 | 0.9500 |
| C13—H13B | 0.9900 | C12'—C13' | 1.395 (2) |
| O14—C15 | 1.3672 (18) | C12'—H12 | 0.9500 |
| C15—C16 | 1.395 (2) | C13'—H13 | 0.9500 |
| C15—C20 | 1.402 (2) | C14'—O1' | 1.2279 (17) |
| C16—C17 | 1.386 (2) | C14'—O2' | 1.3578 (17) |
| C16—H16 | 0.9500 | O2'—C15' | 1.448 (2) |
| C17—C18 | 1.381 (2) | C15'—C16' | 1.482 (3) |
| C17—H17 | 0.9500 | C15'—H15A | 0.9900 |
| C18—C19 | 1.394 (2) | C15'—H15B | 0.9900 |
| C18—H18 | 0.9500 | C16'—H16A | 0.9800 |
| C19—C20 | 1.394 (2) | C16'—H16B | 0.9800 |
| C19—H19 | 0.9500 | C16'—H16C | 0.9800 |
| C20—C21 | 1.4912 (18) | ||
| N24—C1—N25 | 106.41 (11) | N25—C21—C20 | 117.99 (11) |
| N24—C1—C2 | 110.67 (12) | C21—C22—N1' | 120.23 (12) |
| N25—C1—C2 | 110.68 (11) | C21—C22—C23 | 120.64 (12) |
| N24—C1—H1 | 109.7 | N1'—C22—C23 | 119.09 (12) |
| N25—C1—H1 | 109.7 | N24—C23—C4' | 123.99 (12) |
| C2—C1—H1 | 109.7 | N24—C23—C22 | 114.94 (12) |
| C3—C2—C7 | 118.32 (12) | C4'—C23—C22 | 120.95 (12) |
| C3—C2—C1 | 121.92 (13) | C23—N24—C1 | 117.85 (12) |
| C7—C2—C1 | 119.74 (13) | C23—N24—H24 | 115.7 (11) |
| C2—C3—C4 | 121.69 (14) | C1—N24—H24 | 117.0 (11) |
| C2—C3—H3 | 119.2 | C21—N25—C1 | 114.25 (11) |
| C4—C3—H3 | 119.2 | C21—N25—H25 | 112.2 (11) |
| C5—C4—C3 | 119.02 (15) | C1—N25—H25 | 113.9 (11) |
| C5—C4—H4 | 120.5 | C22—N1'—C6' | 110.52 (11) |
| C3—C4—H4 | 120.5 | C22—N1'—C7' | 111.12 (11) |
| C4—C5—C6 | 120.91 (13) | C6'—N1'—C7' | 113.80 (11) |
| C4—C5—H5 | 119.5 | C23—C4'—C14' | 120.22 (12) |
| C6—C5—H5 | 119.5 | C23—C4'—C5' | 120.30 (12) |
| C5—C6—C7 | 119.57 (14) | C14'—C4'—C5' | 119.46 (12) |
| C5—C6—H6 | 120.2 | C4'—C5'—C6' | 111.30 (11) |
| C7—C6—H6 | 120.2 | C4'—C5'—H5A | 109.4 |
| O8—C7—C6 | 123.56 (14) | C6'—C5'—H5A | 109.4 |
| O8—C7—C2 | 115.95 (12) | C4'—C5'—H5B | 109.4 |
| C6—C7—C2 | 120.49 (14) | C6'—C5'—H5B | 109.4 |
| C7—O8—C9 | 118.22 (11) | H5A—C5'—H5B | 108.0 |
| O8—C9—C10 | 106.41 (12) | N1'—C6'—C5' | 113.37 (12) |
| O8—C9—H9A | 110.4 | N1'—C6'—H6A | 108.9 |
| C10—C9—H9A | 110.4 | C5'—C6'—H6A | 108.9 |
| O8—C9—H9B | 110.4 | N1'—C6'—H6B | 108.9 |
| C10—C9—H9B | 110.4 | C5'—C6'—H6B | 108.9 |
| H9A—C9—H9B | 108.6 | H6A—C6'—H6B | 107.7 |
| O11—C10—C9 | 106.62 (12) | N1'—C7'—C8' | 114.11 (12) |
| O11—C10—H10A | 110.4 | N1'—C7'—H7A | 108.7 |
| C9—C10—H10A | 110.4 | C8'—C7'—H7A | 108.7 |
| O11—C10—H10B | 110.4 | N1'—C7'—H7B | 108.7 |
| C9—C10—H10B | 110.4 | C8'—C7'—H7B | 108.7 |
| H10A—C10—H10B | 108.6 | H7A—C7'—H7B | 107.6 |
| C12—O11—C10 | 114.21 (12) | C13'—C8'—C9' | 118.85 (14) |
| O11—C12—C13 | 107.93 (12) | C13'—C8'—C7' | 121.60 (13) |
| O11—C12—H12A | 110.1 | C9'—C8'—C7' | 119.27 (14) |
| C13—C12—H12A | 110.1 | C10'—C9'—C8' | 120.77 (16) |
| O11—C12—H12B | 110.1 | C10'—C9'—H9 | 119.6 |
| C13—C12—H12B | 110.1 | C8'—C9'—H9 | 119.6 |
| H12A—C12—H12B | 108.4 | C11'—C10'—C9' | 120.05 (17) |
| O14—C13—C12 | 106.55 (12) | C11'—C10'—H10 | 120.0 |
| O14—C13—H13A | 110.4 | C9'—C10'—H10 | 120.0 |
| C12—C13—H13A | 110.4 | C10'—C11'—C12' | 120.14 (16) |
| O14—C13—H13B | 110.4 | C10'—C11'—H11 | 119.9 |
| C12—C13—H13B | 110.4 | C12'—C11'—H11 | 119.9 |
| H13A—C13—H13B | 108.6 | C11'—C12'—C13' | 120.06 (17) |
| C15—O14—C13 | 118.19 (12) | C11'—C12'—H12 | 120.0 |
| O14—C15—C16 | 123.66 (14) | C13'—C12'—H12 | 120.0 |
| O14—C15—C20 | 115.89 (12) | C8'—C13'—C12' | 120.10 (15) |
| C16—C15—C20 | 120.43 (14) | C8'—C13'—H13 | 119.9 |
| C17—C16—C15 | 119.99 (15) | C12'—C13'—H13 | 119.9 |
| C17—C16—H16 | 120.0 | O1'—C14'—O2' | 121.26 (13) |
| C15—C16—H16 | 120.0 | O1'—C14'—C4' | 126.49 (13) |
| C18—C17—C16 | 120.57 (14) | O2'—C14'—C4' | 112.25 (12) |
| C18—C17—H17 | 119.7 | C14'—O2'—C15' | 116.83 (12) |
| C16—C17—H17 | 119.7 | O2'—C15'—C16' | 110.61 (17) |
| C17—C18—C19 | 119.21 (14) | O2'—C15'—H15A | 109.5 |
| C17—C18—H18 | 120.4 | C16'—C15'—H15A | 109.5 |
| C19—C18—H18 | 120.4 | O2'—C15'—H15B | 109.5 |
| C20—C19—C18 | 121.60 (15) | C16'—C15'—H15B | 109.5 |
| C20—C19—H19 | 119.2 | H15A—C15'—H15B | 108.1 |
| C18—C19—H19 | 119.2 | C15'—C16'—H16A | 109.5 |
| C19—C20—C15 | 118.12 (13) | C15'—C16'—H16B | 109.5 |
| C19—C20—C21 | 119.28 (13) | H16A—C16'—H16B | 109.5 |
| C15—C20—C21 | 122.59 (12) | C15'—C16'—H16C | 109.5 |
| C22—C21—N25 | 119.80 (12) | H16A—C16'—H16C | 109.5 |
| C22—C21—C20 | 121.84 (12) | H16B—C16'—H16C | 109.5 |
| N24—C1—C2—C3 | 30.99 (17) | C21—C22—C23—N24 | 6.9 (2) |
| N25—C1—C2—C3 | −86.73 (16) | N1'—C22—C23—N24 | −175.27 (12) |
| N24—C1—C2—C7 | −150.56 (13) | C21—C22—C23—C4' | −169.29 (14) |
| N25—C1—C2—C7 | 91.72 (15) | N1'—C22—C23—C4' | 8.5 (2) |
| C7—C2—C3—C4 | −0.4 (2) | C4'—C23—N24—C1 | −157.44 (13) |
| C1—C2—C3—C4 | 178.09 (13) | C22—C23—N24—C1 | 26.47 (18) |
| C2—C3—C4—C5 | 0.0 (2) | N25—C1—N24—C23 | −55.37 (16) |
| C3—C4—C5—C6 | 0.6 (2) | C2—C1—N24—C23 | −175.67 (12) |
| C4—C5—C6—C7 | −0.8 (2) | C22—C21—N25—C1 | −24.73 (19) |
| C5—C6—C7—O8 | −179.74 (15) | C20—C21—N25—C1 | 162.10 (13) |
| C5—C6—C7—C2 | 0.4 (2) | N24—C1—N25—C21 | 53.10 (16) |
| C3—C2—C7—O8 | −179.70 (13) | C2—C1—N25—C21 | 173.39 (12) |
| C1—C2—C7—O8 | 1.80 (18) | C21—C22—N1'—C6' | 139.13 (14) |
| C3—C2—C7—C6 | 0.1 (2) | C23—C22—N1'—C6' | −38.69 (17) |
| C1—C2—C7—C6 | −178.37 (13) | C21—C22—N1'—C7' | −93.54 (16) |
| C6—C7—O8—C9 | 19.4 (2) | C23—C22—N1'—C7' | 88.65 (16) |
| C2—C7—O8—C9 | −160.75 (13) | N24—C23—C4'—C14' | 6.2 (2) |
| C7—O8—C9—C10 | 160.67 (13) | C22—C23—C4'—C14' | −177.91 (13) |
| O8—C9—C10—O11 | −68.86 (15) | N24—C23—C4'—C5' | −172.46 (14) |
| C9—C10—O11—C12 | 163.21 (13) | C22—C23—C4'—C5' | 3.4 (2) |
| C10—O11—C12—C13 | −167.25 (13) | C23—C4'—C5'—C6' | 15.1 (2) |
| O11—C12—C13—O14 | 62.76 (15) | C14'—C4'—C5'—C6' | −163.57 (13) |
| C12—C13—O14—C15 | 172.76 (12) | C22—N1'—C6'—C5' | 57.81 (15) |
| C13—O14—C15—C16 | 0.2 (2) | C7'—N1'—C6'—C5' | −68.03 (15) |
| C13—O14—C15—C20 | −177.99 (12) | C4'—C5'—C6'—N1' | −46.07 (17) |
| O14—C15—C16—C17 | −175.38 (15) | C22—N1'—C7'—C8' | 158.26 (13) |
| C20—C15—C16—C17 | 2.7 (2) | C6'—N1'—C7'—C8' | −76.22 (16) |
| C15—C16—C17—C18 | −0.7 (2) | N1'—C7'—C8'—C13' | −34.0 (2) |
| C16—C17—C18—C19 | −1.4 (2) | N1'—C7'—C8'—C9' | 152.10 (14) |
| C17—C18—C19—C20 | 1.4 (2) | C13'—C8'—C9'—C10' | −1.3 (2) |
| C18—C19—C20—C15 | 0.7 (2) | C7'—C8'—C9'—C10' | 172.83 (16) |
| C18—C19—C20—C21 | 179.96 (13) | C8'—C9'—C10'—C11' | 1.2 (3) |
| O14—C15—C20—C19 | 175.56 (13) | C9'—C10'—C11'—C12' | −0.3 (3) |
| C16—C15—C20—C19 | −2.7 (2) | C10'—C11'—C12'—C13' | −0.6 (3) |
| O14—C15—C20—C21 | −3.7 (2) | C9'—C8'—C13'—C12' | 0.4 (2) |
| C16—C15—C20—C21 | 178.04 (13) | C7'—C8'—C13'—C12' | −173.59 (16) |
| C19—C20—C21—C22 | −49.5 (2) | C11'—C12'—C13'—C8' | 0.6 (3) |
| C15—C20—C21—C22 | 129.79 (16) | C23—C4'—C14'—O1' | −4.2 (2) |
| C19—C20—C21—N25 | 123.54 (15) | C5'—C4'—C14'—O1' | 174.51 (14) |
| C15—C20—C21—N25 | −57.19 (19) | C23—C4'—C14'—O2' | 175.85 (13) |
| N25—C21—C22—N1' | 174.95 (13) | C5'—C4'—C14'—O2' | −5.5 (2) |
| C20—C21—C22—N1' | −12.2 (2) | O1'—C14'—O2'—C15' | 3.5 (2) |
| N25—C21—C22—C23 | −7.3 (2) | C4'—C14'—O2'—C15' | −176.51 (15) |
| C20—C21—C22—C23 | 165.62 (13) | C14'—O2'—C15'—C16' | 88.9 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N24—H24···O1′ | 0.882 (18) | 2.015 (18) | 2.6928 (16) | 132.8 (15) |
| N25—H25···O14 | 0.882 (18) | 2.441 (17) | 2.9744 (17) | 119.3 (13) |
| C6—H6···O1′i | 0.95 | 2.42 | 3.3516 (19) | 168 |
| C18—H18···O1′ii | 0.95 | 2.42 | 3.3613 (19) | 174 |
Symmetry codes: (i) −x, y+1/2, −z+2; (ii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5393).
References
- Anh, L. T., Hieu, T. H., Soldatenkov, A. T., Kolyadina, N. M. & Khrustalev, V. N. (2012b). Acta Cryst. E68, o1588–o1589. [DOI] [PMC free article] [PubMed]
- Anh, L. T., Hieu, T. H., Soldatenkov, A. T., Kolyadina, N. M. & Khrustalev, V. N. (2012c). Acta Cryst. E68, o2165–o2166. [DOI] [PMC free article] [PubMed]
- Anh, L. T., Hieu, T. H., Soldatenkov, A. T., Soldatova, S. A. & Khrustalev, V. N. (2012a). Acta Cryst. E68, o1386–o1387. [DOI] [PMC free article] [PubMed]
- Anh, L. T., Levov, A. N., Soldatenkov, A. T., Gruzdev, R. D. & Hieu, T. H. (2008). Russ. J. Org. Chem. 44, 463–465.
- Bradshaw, J. S. & Izatt, R. M. (1997). Acc. Chem. Res. 30, 338–345.
- Bruker (2001). SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Gokel, G. W. & Murillo, O. (1996). Acc. Chem. Res. 29, 425–432.
- Hieu, T. H., Anh, L. T., Levov, A. N., Nikitina, E. V. & Soldatenkov, A. T. (2009). Chem. Heterocycl. Compd, 45, 1406–1407.
- Hieu, T. H., Anh, L. T., Soldatenkov, A. T., Golovtsov, N. I. & Soldatova, S. A. (2011). Chem. Heterocycl. Compd, 47, 1307–1308.
- Hieu, T. H., Anh, L. T., Soldatenkov, A. T., Kolyadina, N. M. & Khrustalev, V. N. (2012a). Acta Cryst. E68, o2431–o2432. [DOI] [PMC free article] [PubMed]
- Hieu, T. H., Anh, L. T., Soldatenkov, A. T., Kurilkin, V. V. & Khrustalev, V. N. (2012b). Acta Cryst. E68, o2848–o2849. [DOI] [PMC free article] [PubMed]
- Hiraoka, M. (1982). In Crown Compounds. Their Characteristic and Application Tokyo: Kodansha.
- Khieu, T. H., Soldatenkov, A. T., Anh, L. T., Levov, A. N., Smol’yakov, A. F., Khrustalev, V. N. & Antipin, M. Yu. (2011). Russ. J. Org. Chem. 47, 766–770.
- Komarova, A. I., Levov, A. N., Soldatenkov, A. T. & Soldatova, S. A. (2008). Chem. Heterocycl. Compd, 44, 624–625.
- Levov, A. N., Komarova, A. I., Soldatenkov, A. T., Avramenko, G. V., Soldatova, S. A. & Khrustalev, V. N. (2008). Russ. J. Org. Chem. 44, 1665–1670.
- Levov, A. N., Strokina, V. M., Komarova, A. I., Anh, L. T., Soldatenkov, A. T. & Khrustalev, V. N. (2006). Mendeleev Commun. pp. 35–37.
- Pedersen, C. J. (1988). Angew. Chem. Int. Ed. Engl. 27, 1053–1083.
- Sheldrick, G. M. (2003). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sokol, V. I., Kolyadina, N. M., Kvartalov, V. B., Sergienko, V. S., Soldatenkov, A. T. & Davydov, V. V. (2011). Russ. Chem. Bull. 60, 2086–2088.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813007241/cv5393sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813007241/cv5393Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813007241/cv5393Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




