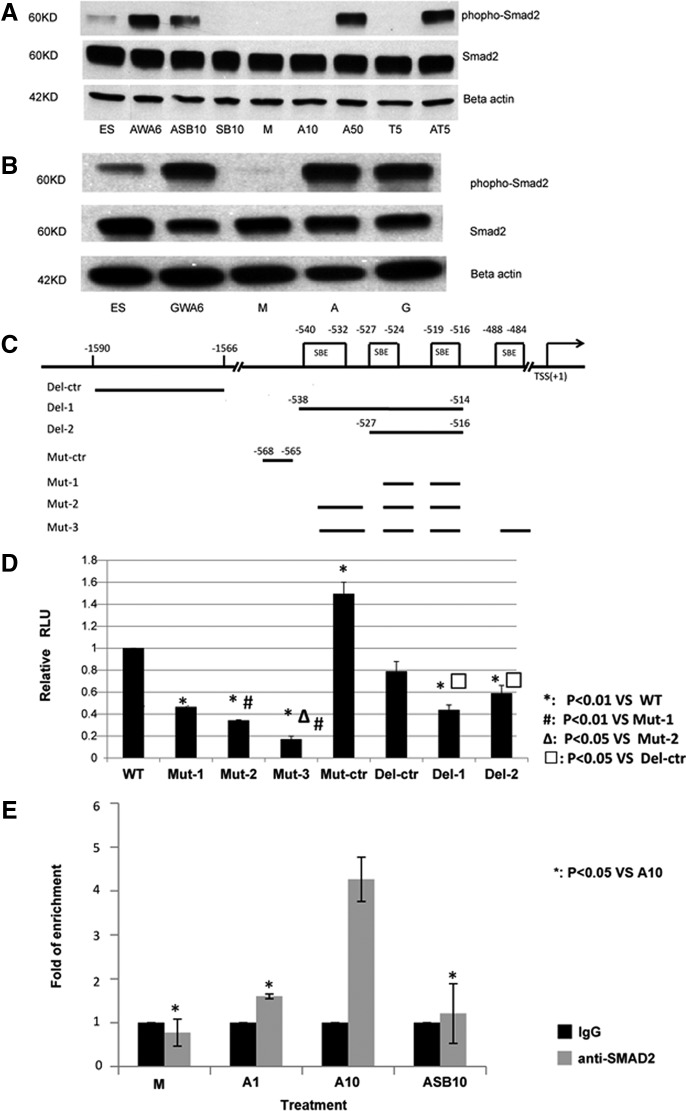
FIG. 4.
NKX2-1 expression by the Activin-A is dependent on SMAD2 activation. (A–C) Western blot analysis of phospho-SMAD2 and total SMAD2 in whole-cell lysates. (A) Undifferentiated hESC-H9=ES; hESC-H9 differentiated with Activin-A+Wnt3a d0–d4 and Activin-A between d4–6=AWA6; hESC-H9 differentiated between d6–d8 with Activin-A+SB431542 (ASB10), SB431542 alone (SB10), medium (M), 10 ng/mL Activin-A (A10), 50 ng/mL Activin-A (A50), 5 ng/mL TGFβ1 (T5), Activin-A+TGFβ1 (AT5). (B) Undifferentiated hESC-H9=ES; hESC-H9 differentiated with 250 ng/mL GDF11+Wnt3a between d0–d4 and GDF11 between d4–6 (GWG6); between d6–d8 with medium (M), 50 ng/mL Activin-A (A) or 250 ng/mL GDF11 (G). (C) Schematic representation of NKX2-1 distal promoter region (not to scale). TSS: transcription start site; SBE: SMAD binding element. (D) NCI-H441 cells were transfected with wild-type pNKX2-1-luc(WT), pNKX2-1(mSBE1)-Luc(Mut-1), pNKX2-1(mSBE2)-Luc(Mut-2), pNKX2-1(mSBE3)-Luc(Mut-3), pNKX2-1(mSBEctr)-Luc(Mut-ctr), pNKX2-1(dSBE1)-Luc(Del-1), pNKX2-1(dSBE2)-Luc(Del-2), pNKX2-1(dSBEctr)-Luc(Del-ctr), on day 1 (all with pGV). Cells were harvested after 2 days of treatment and luciferase activity was analyzed. (*: P<0.01 vs. WT; #: P<0.01 vs. Mut-1; Δ: P<0.05 vs. Mut-2; □: P<0.05 vs. Del-ctr). (E) chromatin immunoprecipitation (ChIP) assay for SMAD binding to the NKX2-1 distal promoter. Differentiated hESC-H9 cells were cultured in M, 10 ng/mL Activin-A, 100 ng/mL Activin-A, or 100 ng/mL Activin-A+10 μM SB431542 for another 2 days. Cells were harvested and ChIP performed with an isogenic or anti-SMAD2 antibodies. Enrichment for the NKX2-1 promoter in the precipitated DNA was analyzed by qPCR using primers that flank the distal promoter region. Results from triplicate experiments are shown as fold change of DNA enrichment. *P<0.05.