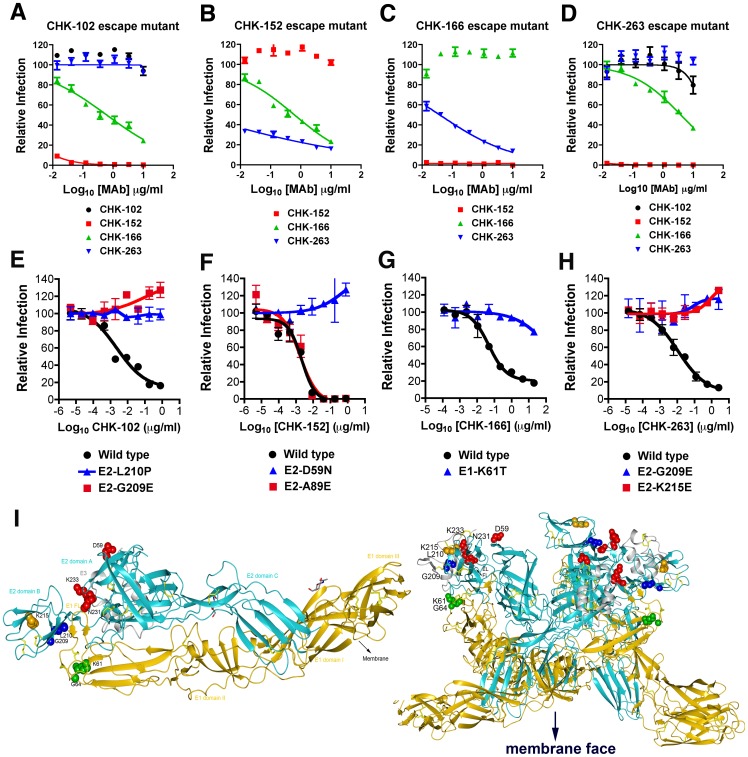
Figure 6. Characterization and mapping of neutralization escape mutants.
A–D. FRNT assay with bulk virus obtained after three to six passages under selection of (A) CHK-102, (B) CHK-152, (C) CHK-166, or (D) CHK-263 on Vero cells. Bulk virus also was tested for infectivity in the presence of the non-selecting MAbs. Results are representative of two to three independent experiments performed in triplicate. E–H. Confirmation of resistant phenotype with SFV-CHIKV-GFP containing the indicated single engineered point mutations. Serial dilutions of MAb were incubated with chimeric SFV-CHIKV virus (WT or mutant stocks) for one hour at room temperature. MAb-virus complexes were added to Vero cells plated in 96-well plates and incubated at 37°C. After 8 hours cells were trypsinized, fixed, and the number of GFP-positive, infected cells was assessed by flow cytometry. Curves are representative of 2 to 3 independent experiments. I. Epitope mapping of anti-CHIKV MAbs on the crystal structure of the mature envelope glycoprotein complex (PDB code 3N44). (Left) The domains on E2 (cyan) and E1 (gold) are indicated, and the fusion loop on E1 (E1 FL) is delineated. Amino acid residues of neutralizing MAbs were determined by escape selection, sequencing, and reverse genetic confirmation. CHK-102 and CHK-263 recognize the B domain on E2, CHK-152 recognizes a residue on the wings of the A domain on E2, and CHK-166 recognizes an amino acid in domain II of E1 proximal to the conserved fusion loop. (Right) The mature envelope glycoprotein docked onto the trimer conformation (PDB code 2XFB) that is present on the virion. E3, E2, and E1 and the escape residues are colored as in the left panel. Neutralization escape residues are readily accessible on the top of the trimer, distal to the viral membrane.