Abstract
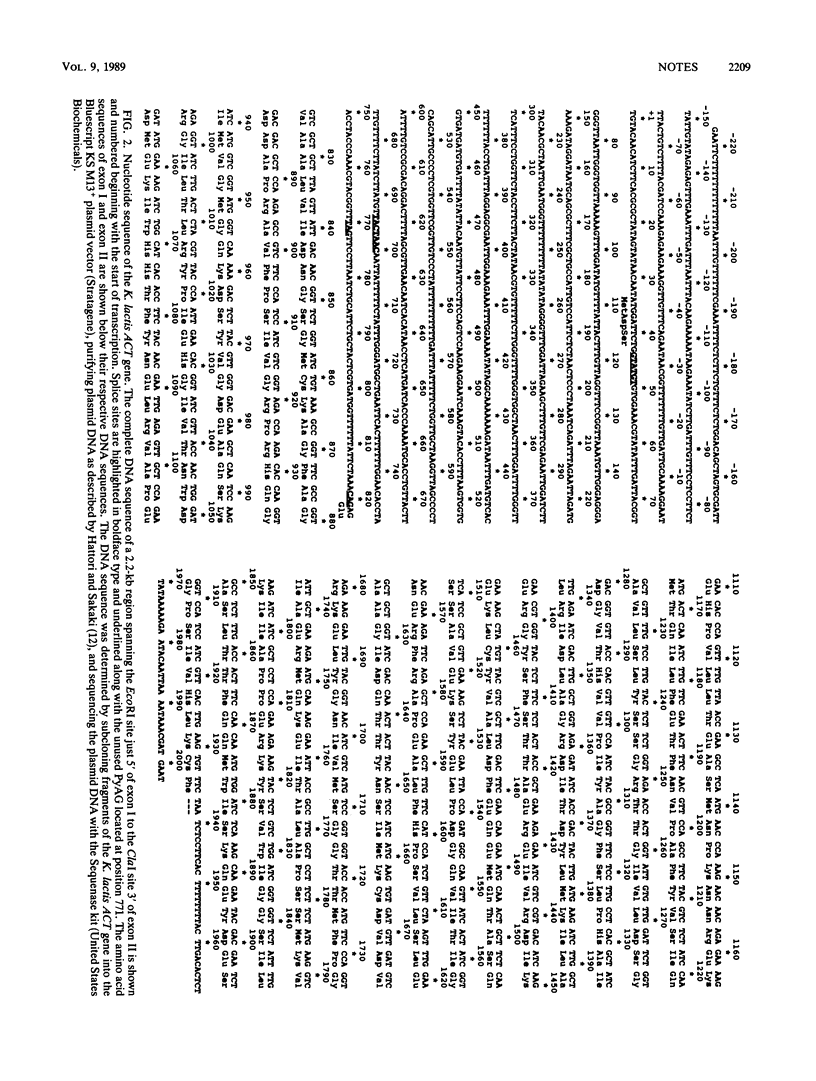
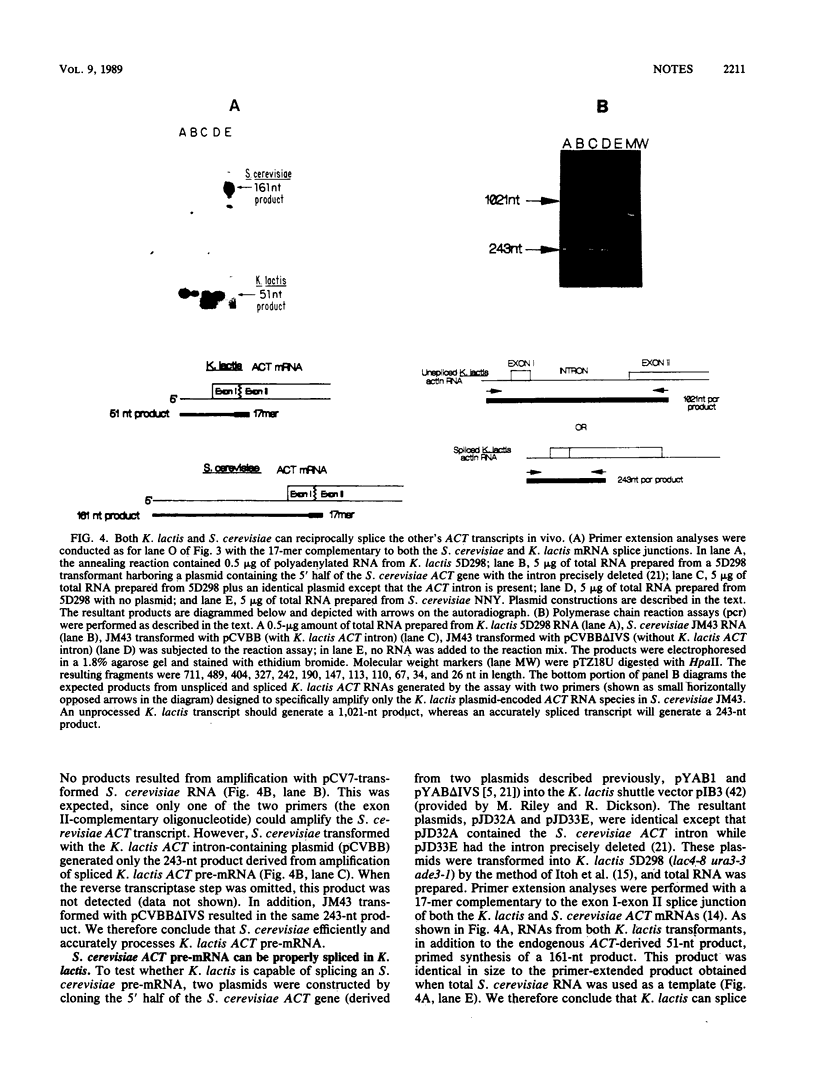
The actin (ACT) gene from the budding yeast Kluyveromyces lactis was cloned, and the nucleotide sequence was determined. The gene had a single intron 778 nucleotides in length which possessed the highly conserved splicing signals found in Saccharomyces cerevisiae introns. We demonstrated splicing of heterologous ACT transcripts in both K. lactis and S. cerevisiae.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986 Oct 10;47(1):49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- Beggs J. D., van den Berg J., van Ooyen A., Weissmann C. Abnormal expression of chromosomal rabbit beta-globin gene in Saccharomyces cerevisiae. Nature. 1980 Feb 28;283(5750):835–840. doi: 10.1038/283835a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cellini A., Felder E., Rossi J. J. Yeast pre-messenger RNA splicing efficiency depends on critical spacing requirements between the branch point and 3' splice site. EMBO J. 1986 May;5(5):1023–1030. doi: 10.1002/j.1460-2075.1986.tb04317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellini A., Parker R., McMahon J., Guthrie C., Rossi J. Activation of a cryptic TACTAAC box in the Saccharomyces cerevisiae actin intron. Mol Cell Biol. 1986 May;6(5):1571–1578. doi: 10.1128/mcb.6.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Physiological studies of beta-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980 Jun;142(3):777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Fouser L. A., Friesen J. D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986 Apr 11;45(1):81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- Gallwitz D. Construction of a yeast actin gene intron deletion mutant that is defective in splicing and leads to the accumulation of precursor RNA in transformed yeast cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3493–3497. doi: 10.1073/pnas.79.11.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Perrin F., Seidel R. The actin gene in yeast Saccharomyces cerevisiae: 5' and 3' end mapping, flanking and putative regulatory sequences. Nucleic Acids Res. 1981 Dec 11;9(23):6339–6350. doi: 10.1093/nar/9.23.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Herman A., Roman H. Allele specific determinants of homothallism in Saccharomyces lactis. Genetics. 1966 Apr;53(4):727–740. doi: 10.1093/genetics/53.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Rymond B. C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987 Aug 14;50(4):593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Langford C., Nellen W., Niessing J., Gallwitz D. Yeast is unable to excise foreign intervening sequences from hybrid gene transcripts. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1496–1500. doi: 10.1073/pnas.80.6.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G. P., Itakura K., Ito H., Rossi J. J. Saccharomyces cerevisiae actin--Escherichia coli lacZ gene fusions: synthetic-oligonucleotide-mediated deletion of the 309 base pair intervening sequence in the actin gene. Gene. 1983 Apr;22(1):31–39. doi: 10.1016/0378-1119(83)90061-6. [DOI] [PubMed] [Google Scholar]
- Mertins P., Gallwitz D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J. 1987 Jun;6(6):1757–1763. doi: 10.1002/j.1460-2075.1987.tb02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa G. J., Zaia J. A., Spallone P. A., Stephens D. A., Kaplan B. E., Wallace R. B., Rossi J. J. Direct detection of HIV-1 RNA from AIDS and ARC patient samples. DNA. 1988 May;7(4):287–295. doi: 10.1089/dna.1988.7.287. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987 Dec 1;6(12):3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987 Jun 5;49(5):613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Riedel N., Wolin S., Guthrie C. A subset of yeast snRNA's contains functional binding sites for the highly conserved Sm antigen. Science. 1987 Jan 16;235(4786):328–331. doi: 10.1126/science.2948278. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986 Nov;6(11):3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Jones M. H., Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987 Sep 18;237(4821):1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986 Jul;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]