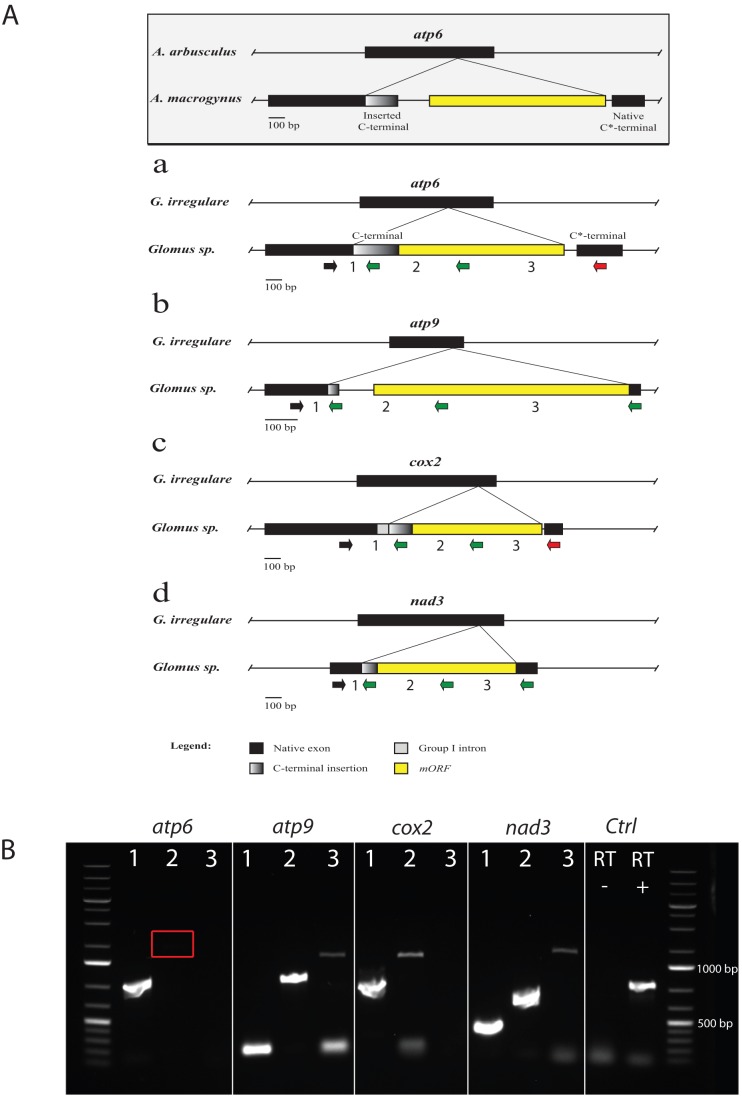
Figure 4. Comparison of gene hybrids atp6, atp9, cox2 and nad3.
A) The atp6 gene hybrid reported for Allomyces macrogynus (grayscale, boxed) is used as a reference in a comparison of the most similar atp6 (a), atp9 (b), cox2 (c) and nad3 (d) genes in Glomus sp. mtDNA. Each occurrence is put in perspective with the gene of a close relative (either Allomyces arbusculus or G. irregulare) in order to show the insertion point of the foreign element with the projections. Exons are in black, while the inserted foreign C-terminal is shaded in gray. The mobile endonuclease element is in yellow. For each gene, the black arrow indicates the position of the forward primer used in the downstream RT-PCR experiment in combination with three different reverse primers. The green arrows indicate expression at the RNA level of the corresponding portion of the gene, while the red arrows indicate a negative amplification. B) Agarose gel electrophoresis figure showing the RT-PCR results. For each gene hybrid, the expression at the RNA level was tested using a forward primer in the conserved gene core and a reverse primer respectively in the inserted C-terminal (1), the mobile endonuclease (2) and the native C*-terminal portion (3). Primers in nad5 exon 4 were used as a positive control on cDNA (RT +) and negative control on RNA (RT −). The expected size of the amplified fragments was: atp6 inserted C-terminal (684 bp), atp9 inserted C-terminal (149 bp), atp9 mORF (717 bp), atp9 native C*-terminal (1085 bp), cox2 inserted C-terminal (938 bp), cox2 mORF (1291 bp), nad3 inserted C-terminal (261 bp), nad3 mORF (597 bp), nad3 native C*-terminal (1183 bp) and the positive control in nad5 exon 4 had an expected amplicon size of 689 bp. The red box indicates a faint band that is present on the gel.