Figure 5. Residues that form contacts early in the folding (pre-TS) and constitute aggregation-prone regions in CRABP1 are evolutionarily conserved in the iLBP family.
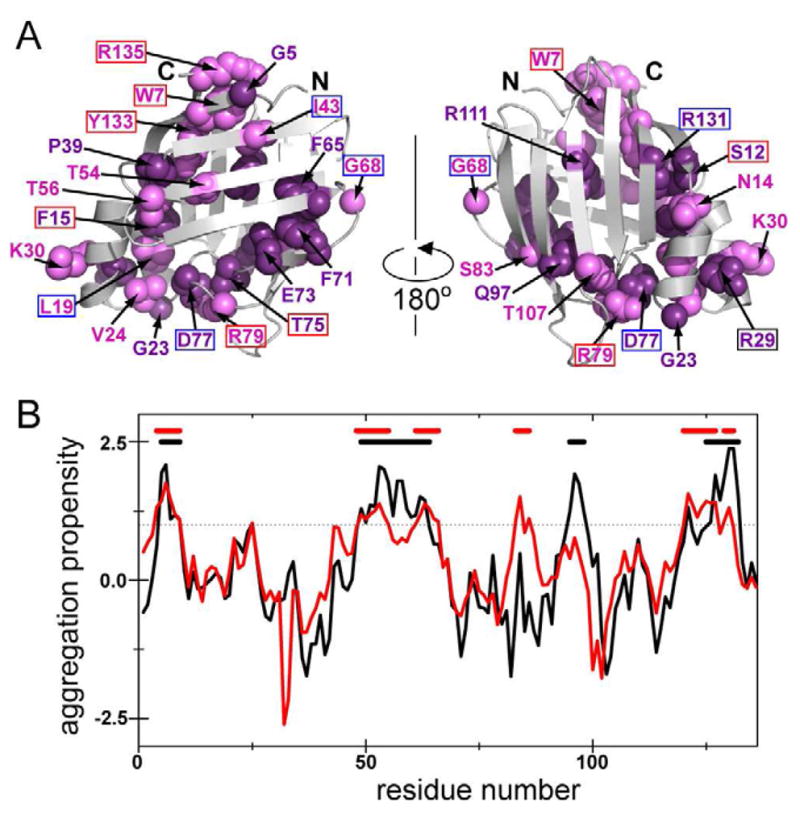
(A) Sequence conservation residues in the iLBP family. Multiple sequence alignment of iLBPs to CRABP1 was carried out using the ConSurf server (Ashkenazy et al., 2010). Residues shown in dark purple spheres on the CRABP1 structure have the highest ConSurf score (9) and those shown in light purple have the next highest (8). Residue numbering corresponds to that in the CRABP1 sequence. Boxed residues were subjected to perturbation analysis in this study, and the box color corresponds to φ-values (red for φ > 0.5 and blue for φ < 0.5).
(B) Aggregation propensity profiles for CRABP1 and iLBP family average predicted by Zyggregator (Tartaglia and Vendruscolo, 2008). The aggregation propensity profile for CRABP1 WT is shown in black, and the average iLBP aggregation profile in red. The horizontal dotted line represents the 1.0 threshold for significant aggregation propensity; continuous stretches of residues with aggregation score above the threshold are considered aggregation-prone and marked as black and red horizontal bars, respectively.
