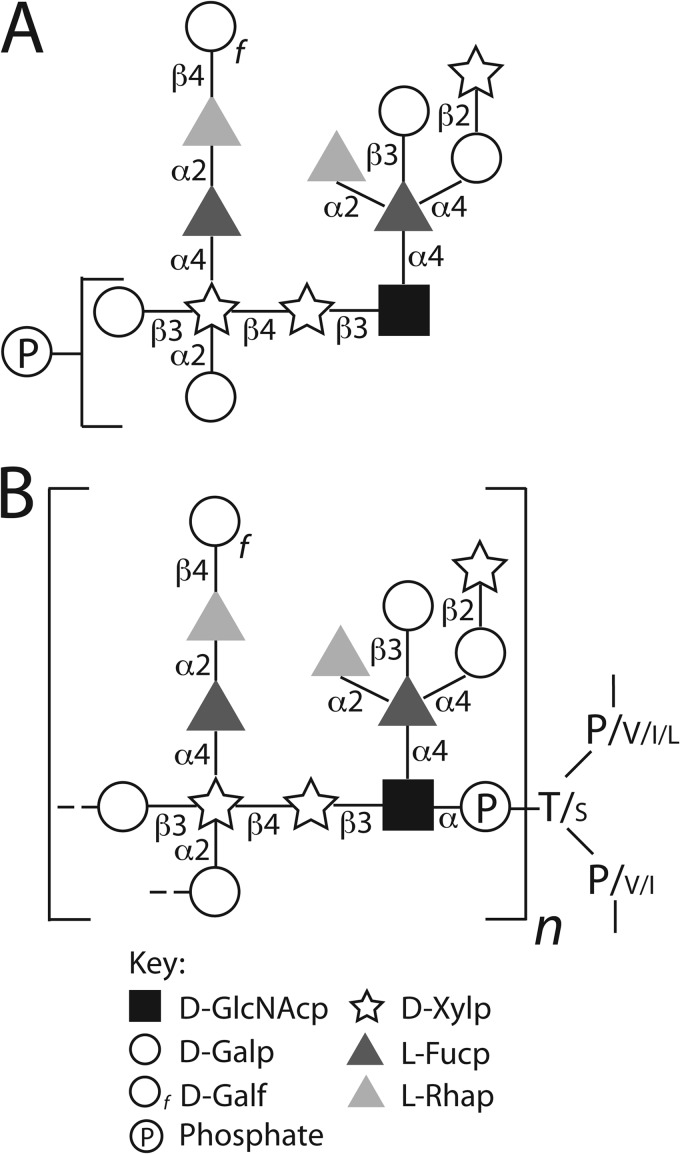
FIGURE 7.
The proposed fine structure of the WIC29.26 phosphosaccharide epitope found on the gp72 glycoprotein of T. cruzi epimastigotes. A, the chemical structure of the largest phosphosaccharide structure released from gp72 Pronase glycopeptides by partial aq. HF dephosphorylation. The precise location of the phosphate group (found in both phosphomonoester and cyclic phosphodiester form) is unknown but is linked to one or other of the two terminal Galp residues, as indicated. The heterogeneity of the structures observed by ES-MS and NMR suggests that not every phosphosaccharide unit is complete. B, proposed arrangement of the phosphosaccharide units on gp72. The proposed repeating pattern of n phosphosacchride units (with putative GlcNAcα1-P-Gal inter-repeat linkages) is based on the ratio of phosphosacchride units to potential phosphoglycosylation sites (30:18), suggested by compositional analyses of gp72 (13, 14) and its amino acid sequence (15), and is supported by the position of the phosphate group shown in A. The phosphodiester linkage to Thr (T) and Ser (S) is based on Ref. 11, and the sizes of the amino acid codes reflect their relative frequencies in the amino acid sequences of the putative gp72 phosphoglycosylation sites (15). The symbols are as defined in the legend to Fig. 4.