FIGURE 4.
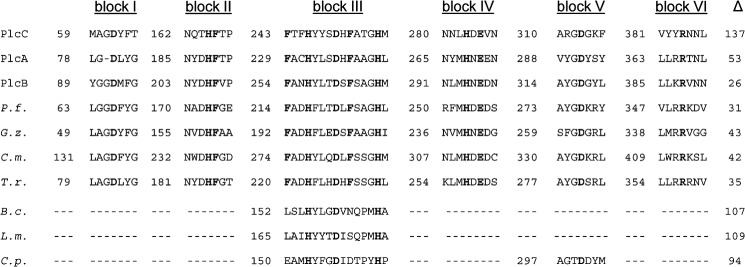
Partial amino acid sequence alignment of L. pneumophila PLC enzymes with P. fluorescens PlcC, including uncharacterized homologous proteins and protein domains/motifs essential for PLC activity. Upper panel, partial amino acid sequence alignment of L. pneumophila PlcC, PlcA, and PlcB with P. fluorescens PlcC (P.f., gi 11611251) and uncharacterized homologs of different pathogenic fungi: G. zeae (G.z., gi 46119808), C. militaris CM01 (C.m., gi 346326200), and T. rubrum (T.r., gi 327307586). Lower panel, homologous regions from characterized PLC enzymes of different bacterial species: B. cereus (B.c., gi 28414376), L. monocytogenes (L.m., gi 374922439), and C. perfringens C.p., gi 110675499). Experimentally determined essential amino acids for PlcC activity are shown in bold. Numerals indicate the number of amino acids before the start of the homologous region. The Δ column designates the length of the C-terminal domain proceeding after the last homology region.
