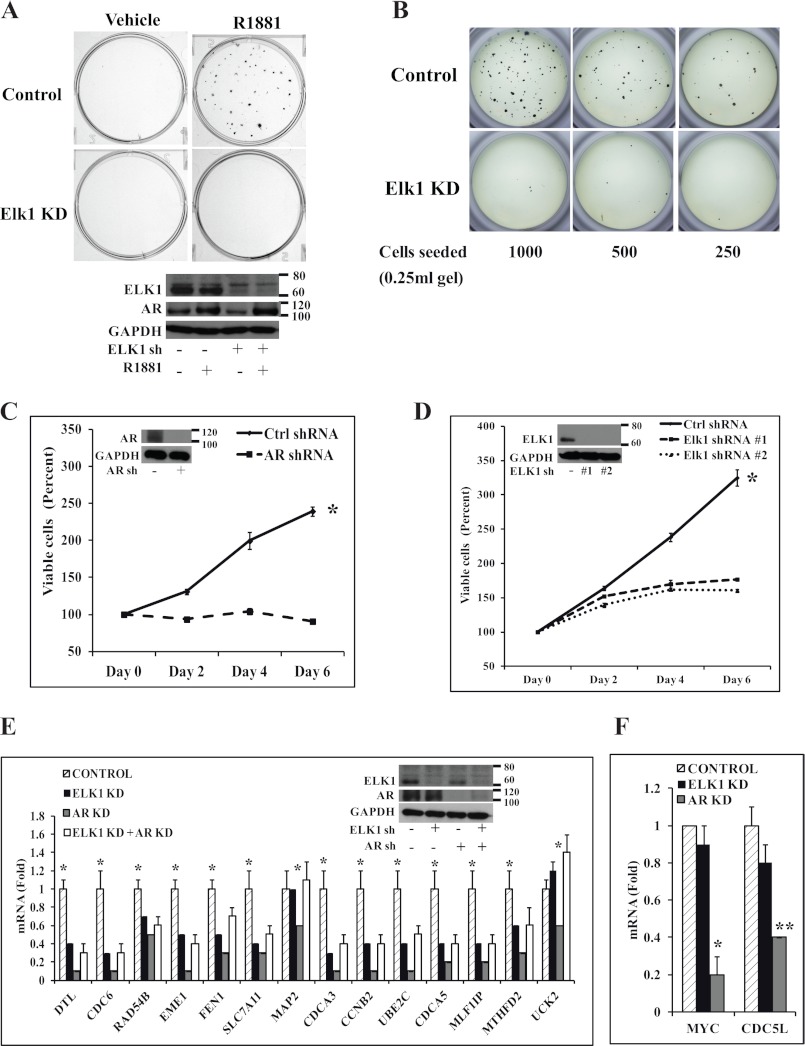
FIGURE 3.
Effect of depleting ELK1 on clonogenic survival, growth, and AR-regulated gene expression in CRPC cells. A, top, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), anchorage-dependent colony formation was measured following treatment with vehicle or R1881 (1 nm). At the end of 2 weeks, colonies were stained with crystal violet. A, bottom, at the end of 48 h of treatment, the cells were harvested for Western blot to probe for ELK1 and AR with GAPDH as the loading control. B, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), anchorage-independent colony formation was measured in 0.8% soft agar containing a serial dilution of cells following treatment with R1881 (1 nm). At the end of 2 weeks, colonies were stained with MTT. C, hormone-depleted C4-2 cells were infected with AR shRNA lentivirus or control (Ctrl) shRNA lentivirus. 72 h later (after the knockdown had occurred), cell growth was monitored in hormone-depleted medium by the MTT assay. C, inset, Western blot showing AR with GAPDH as the loading control. D, hormone-depleted C4-2 cells were infected with ELK1 shRNA 1, ELK1 shRNA 2 lentivirus, or control shRNA lentivirus. 72 h later (after the knockdown had occurred), cell growth was monitored in hormone-depleted medium by the MTT assay. D, inset, Western blot showing ELK1 with GAPDH as the loading control. E, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA, AR shRNA lentivirus, a combination of AR shRNA and ELK1 shRNA lentivirus, or control shRNA lentivirus. 72 h after infection, cells were seeded in 6-well plates and incubated for an additional 72 h to achieve a good combined knockdown of ELK1 and AR. Cells were harvested to measure mRNA levels for the indicated genes by quantitative real time PCR. E, inset, Western blot showing ELK1 and AR with GAPDH as the loading control. F, the RNA samples used in E were analyzed for expression of two previously known apo-AR-regulated genes by quantitative real time PCR. KD denotes knockdown. *, p < 0.001; **, p < 0.001. Error bars represent S.D.