Figure 1.
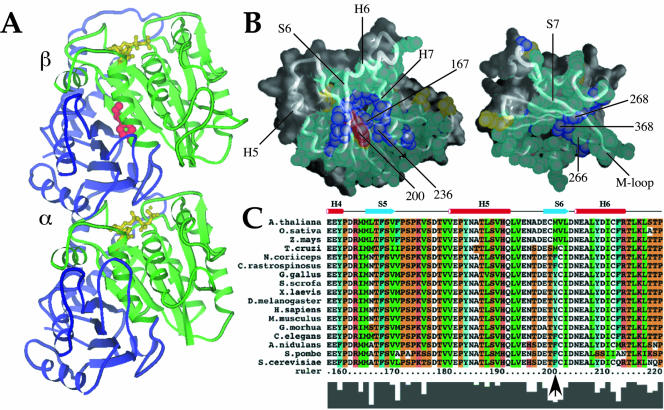
A conserved internal residue corresponding to position 200 in mammalian β-tubulin is positioned to affect interdomain motion. (A and B) Diagram of α/β tubulin dimer from the crystal structure database. Green, nucleotide binding domain comprising residues 1–240; red, residue 200; yellow, nucleotide; blue, remainder of protein. (B) Contacts made between the nucleotide-binding (left) and intermediate (right) domains of porcine β-tubulin. Atoms in residues involved in contacts are shown as spheres. Backbone worm is superimposed to indicate secondary structure with selected helices and sheets labeled for orientation. Residues colored in aqua are highly conserved among all known β-tubulins (126 sequences from Swiss Prot); purple residues show only conservative substitutions; yellow residues show nonconservative substitutions. Residue 200 falls into this last class and is colored red for distinction. Numbered atoms are those implicated in benzimidazole binding. Alpha helices H5, H6, and H7, beta strands S6 and S7, and the M-loop (Löwe et al., 2001) are indicated. (C) Primary β-tubulin sequences from representative eukaryotes showing the immediate region of interest: plants (A. thalania, Z. mays, O. Sativa), human (H. sapiens), mouse (M. musculus), chicken (G. gallus), pig (S. scrofa), frog (X. laevis), fly (D. melanogaster), psychrotolerant fish (G. morhua), worm (C. elegans), psychrophilic fish (N. coriiceps, C. rastrospinosus), and fungi (S. cerevisiae, A. nidulans, S. pombe). Numbering is according to human β-tubulin. The conserved phyla-specific residue corresponds to human Y200, Antarctic fish F200, and S. pombe F200 and is indicated by an arrow. Residues are colored as follows: orange, G, P, S, and T; red, H, K, and R; blue, W and Y; aqua, F; green, I, L, M, and V; and white, A, C, N, D, E, and Q. The histogram at the bottom of the sequences reflects conservation of amino acids at each position.