A DOCK2 mutation in established Irf5 −/− mice has specific effects on B cells.
Keywords: autoimmunity, Th1, Th2, type I IFN, systemic lupus erythematosus
Abstract
Interferon regulatory factor 5-deficient (IRF5 −/−) mice have been used for many studies of IRF5 biology. A recent report identifies a mutation in dedicator of cytokinesis 2 (DOCK2) as being responsible for the abnormal B-cell development phenotype observed in the IRF5 −/− line. Both dedicator of cytokinesis 2 (DOCK2) and IRF5 play important roles in immune cell function, raising the issue of whether immune effects previously associated with IRF5 are due to IRF5 or DOCK2. Here, we defined the insertion end-point of the DOCK2 mutation and designed a novel PCR to detect the mutation in genomic DNA. We confirmed the association of the DOCK2 mutation and the abnormal B-cell phenotype in our IRF5 −/− line and also established another IRF5 −/− line without the DOCK2 mutation. These two lines were used to compare the role of IRF5 in dendritic cells (DCs) and B cells in the presence or absence of the DOCK2 mutation. IRF5 deficiency reduces IFN-α, IFN-β and IL-6 production by Toll-like receptor 9 (TLR9)- and TLR7-stimulated DCs and reduces TLR7- and TLR9-induced IL-6 production by B cells to a similar extent in the two lines. Importantly however, IRF5 −/− mice with the DOCK2 mutation have higher serum levels of IgG1 and lower levels of IgG2b, IgG2a/c and IgG3 than IRF5 −/− mice without the DOCK2 mutation, suggesting that the DOCK2 mutation confers additional Th2-type effects. Overall, these studies help clarify the function of IRF5 in B cells and DCs in the absence of the DOCK2 mutation. In addition, the PCR described will be useful for other investigators using the IRF5−/− mouse line.
Introduction
Interferon regulatory factor 5 (IRF5) is a member of the IRF family of transcription factors that collectively are involved in the regulation of innate immune responses, immune cell development and oncogenesis (1, 2). IRF5 has a number of functions including the induction of pro-inflammatory cytokines and type I interferons following viral infection or downstream of Toll-like receptors (TLRs), nucleotide-binding oligomerization domain 2 and retinoic acid-inducible gene I (3–8). In addition, IRF5 plays a role in apoptotic pathways induced by viral infection, DNA damage, Fas ligand or tumor necrosis factor-related apoptosis-inducing ligand (7, 9, 10).
IRF5 polymorphisms are associated with an increased risk of developing a number of human autoimmune diseases including systemic lupus erythematosus, scleroderma, rheumatoid arthritis, Sjögren’s syndrome and ulcerative colitis (11–18). These IRF5 polymorphisms cause the expression of novel IRF5 isoforms (11, 12) and/or an increased level of IRF5 expression by promoting the stability of the IRF5 mRNA or protein (19–21). In relation to lupus, interest has focused on the role of IRF5 in TLR signaling as dysregulated TLR7 and TLR9 responses to endogenous RNA and DNA have been linked to lupus pathogenesis (22–26). We have previously found that IRF5 is required for IFN-α, IFN-β and IL-6 production by mouse dendritic cells (DCs) following TLR7 activation by RNA-containing immune complexes and/or TLR7 and TLR9 activation by synthetic ligands (6). A number of studies have shown that IRF5 deficiency reduces disease severity in mouse models of lupus (27–30).
Many of the studies evaluating the role of IRF5 biology including its role in lupus have involved the use of the IRF5 knockout mouse line (5). As part of our ongoing efforts to understand how IRF5 may contribute to lupus pathology we have observed, as shown in this report, that IRF5 −/− mice backcrossed 11 generations to the C57BL/6 genetic background had a marked reduction in the percentage of mature B cells and almost no splenic marginal zone B cells. Surprisingly, this B-cell phenotype was lost in IRF5 −/− mice backcrossed further to C57BL/6, indicating that this B-cell developmental phenotype was not due to IRF5 deficiency per se. A recent report describes a similar B-cell phenotype in IRF5 −/− mice to that which we have found and demonstrates that this is due to a previously unrecognized mutation of the dedicator of cytokinesis 2 (DOCK2) gene in IRF5 −/− mice resulting in reduced expression of DOCK2 (31). This raises the possibility that some effects attributed to IRF5 in previous studies using the IRF5 −/− mice may have been due to the DOCK2 mutation and not to IRF5. This is a particular concern as DOCK2, a hematopoietic cell-specific guanine exchange factor that mediates Rac activation, plays a role in immune responses. DOCK2 −/− mice exhibit migration defects of B lymphocytes, T lymphocytes and neutrophils, due to defective chemokine receptor signaling (32, 33). DOCK2 −/− mice develop excessive T helper cell type 2 (Th2) responses as a result of the failure of DOCK2-deficient CD4+ T cells to down-regulate the expression of surface IL-4 receptor α (34). Furthermore, plasmacytoid dendritic cells (pDCs) from DOCK2 −/− mice have an impaired ability to produce IFN-α and IFN-β in response to TLR7 and TLR9 ligands (35).
In this study, we report a novel PCR that can be used to identify the DOCK2 mutation responsible for the decreased expression of DOCK2. We have used this PCR to identify which mice in our colony express the DOCK2 mutation and find that the abnormal B-cell phenotype is associated with the presence of the DOCK2 mutation, consistent with the recent findings of Purtha et al. (31). We have also compared TLR-induced responses of B cells and DCs from IRF5 −/− mice with and without the DOCK2 mutation to determine the relative contribution of DOCK2 and IRF5 to these responses. In addition, we have compared serum IgG isotype and IgM levels in these mice to determine the extent to which the observed Th2-type IgG isotype skewing observed in the IRF5 −/− line is due to IRF5 deficiency or to the presence of the DOCK2 mutation.
Methods
Mice
C57BL/6 wild-type mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). IRF5 −/− mice backcrossed eight generations to C57BL/6 were provided by Dr T. Taniguchi (University of Tokyo, Tokyo, Japan) with the permission of Dr T. Mak (University of Toronto, Toronto, Canada) (5). The IRF5 −/− mice were further backcrossed using C57BL/6 mice from The Jackson Laboratory to make the 11th, 14th and 15th generation backcrossed mice used in this study. These are termed IRF5 −/− 11G, IRF5 −/− 14G and IRF5 −/− 15G, respectively. All IRF5 −/− 11G mice used in this study had a complete B-cell phenotype abnormality that included an absence of marginal zone B cells. All mice were maintained at the Boston University School of Medicine Laboratory Animal Sciences Center in accordance with the regulations of the American Association for the Accreditation of Laboratory Animal Care. All experimental procedures were approved by the institutional animal care and use committee at Boston University School of Medicine.
Single nucleotide polymorphism analysis
Single nucleotide polymorphisms (SNPs) in genomic DNA were analyzed by the JAX Mouse Diversity Genotyping Array Service (The Jackson Laboratory) using DNA obtained from mouse tails. This analysis measures >500 000 SNPs (approximately every 5kb) in the mouse genome.
Reverse transcriptase PCR
RNA was purified from spleen cells of Jackson C57BL/6 mice, IRF5 −/− 15G mice and IRF5 −/− 11G mice using Trizol (Invitrogen, Grand Island, NY, USA). cDNA was made using the ThermoScript reverse transcriptase (RT)–PCR system for first-strand cDNA synthesis (Invitrogen). PCR using cDNA was performed using GoTaq Flexi DNA polymerase (Promega, Madison, WI, USA) and primers as previously published (Table 1) (31).
Table 1.
PCR primers used in this study
| Primer name | Sequences |
|---|---|
| PCR using cDNA to detect DOCK2 mutation (31) | |
| DOCK2Ex27-30F (forward) | GGA TGC GGC CTT CAC TTA |
| DOCK2Ex27-30R (reverse) | TCC ACA GCT GGA ACT CAA AG |
| DOCK2Ex29-28F (forward) | CAA GGA CCT CAT TGG GAA GAA |
| DOCK2Ex29-28R (reverse) | TCT GAG CTG GTC TGG AAG GTC T |
| Long-range PCR to detect insertion position of the DOCK2 duplication | |
| Ex29F2 (forward) | GAA GAA TGT GTA TCC TGG GGA CTG G |
| R15 (reverse) | ATA AAG CCT GCA GAG ACA GGA AGC AG |
| R16 (reverse) | AGG AGC ATC TTC TCA GGA GCC TTC |
| R17 (reverse) | GAG AGT GAC CCA CAA GAC TGA ATG G |
| R18 (reverse) | CTG TCT GCT CTC CTG GGT AAA TGT C |
| R19 (reverse) | ACT ATG ACT GGA GGG CCT GAA GAT TG |
| R20 (reverse) | ACC TTC AGG GAC CCT TTC TCA CTG |
| R21 (reverse) | ACA GCT GTA CCT GAG CAC CCA TGC |
| R22 (reverse) | GGG TGA TTA GGA GGC TTC TTC TGC |
| R23 (reverse) | CCC TCA TCC CTC CAT TTC TAC AGA G |
| R24 (reverse) | CAG AAT AAG GAC AGA GGC AGA GGT G |
| R25 (reverse) | AGG TCC TGC TGT GTG GTA GAT GAA G |
| R26 (reverse) | GAC CAA AGG CAT CAC TCA GTC TCT C |
| Ex28R2 (reverse) | GAG ACC TTC CAG ACC AGC TCA GAC |
| In29.1F (forward) | GCC TGT ACC ATA CAA AGC CCT TCA TC |
| In29.2F (forward) | GAA GAT CGA AGA AAC CGA CAC CTC TC |
| In29.3F (forward) | CTC AGG GTG CCT AGT GTT ATC ACC TC |
| In29.4F (forward) | GAC CTT ATG AGG TGG AAC CAC AAC C |
| In29.5F (forward) | GTG TGT GTC ATG TAG CTG ACA TCC TG |
| In29.6F (forward) | ACA GAG TAC TTC CAG CCT CAG TTT CC |
| In29.7F (forward) | CAT TCC TAC TGC TGT GTG GTC CTG |
| In29.8F (forward) | GCA CGT CCT TTA TAG GTC TCC TCT GG |
| In29.9F (forward) | AGG AGA AGA AAG CTG TGG GAA CAG TC |
| PCR using genomic DNA to detect DOCK2 mutation | |
| In29.4F (forward) | GAC CTT ATG AGG TGG AAC CAC AAC C |
| InR22.3.1R (reverse) | GAT CCA AAG ATT CCC TAC AGC TCC AC |
| Internal control (CD19) to confirm adequacy of DNA preparation | |
| oIMR1589 | CCT CTC CCT GTC TCC TTC CT |
| oIMR1590 | TGG TCT GAG ACA TTG ACA ATC A |
PCR
To detect the 3′ end of the DOCK2 duplication, genomic DNA was purified from spleen cells of IRF5 −/−11G mice using Archive Pure DNA cell/tissue kit (5 Prime, Gaithersburg, MD, USA). Primers used in this study are listed in Table 1. Long-range PCR was performed using LongAmp Taq DNA polymerase (New England BioLabs, Ipswich, MA, USA), which can amplify up to 30kb. For the final PCR, which detects the DOCK2 mutation as a 305-bp product, DNA was obtained from tails digested with Proteinase K (5 Prime) and PCR was performed using GoTaq Flexi DNA polymerase (Promega). PCR primer sequences that recognize the CD19 gene were obtained from the Web site of The Jackson Laboratory (http://jaxmice.jax.org/strain/004126.html), and the primers used as an internal control to ensure that DNA preparation was adequate in all tail DNA samples.
Reagents
Pam3CSK4, Poly (I:C), LPS, CL097, CpG-A ODN 2336 (ggg GAC GAC GTC GTG ggg ggg) and CpG-B ODN 1826 (tcc atg acg ttc ctg acg tt) were purchased from InvivoGen (San Diego, CA, USA). Capital letters in ODN sequence indicate a phosphodiester backbone, and lowercase letters indicate a phosphorothioate backbone. The goat anti-mouse IgM antibody [F(ab’)2 fragment; µ chain specific] was obtained from Jackson ImmunoResearch (West Grove, PA, USA).
Staining of spleen cells and bone marrow cells
Spleen cell suspensions and bone marrow cells were treated with red blood cell lysing buffer (Sigma, St Louis, MO, USA) to remove red blood cells. The spleen cells were stained with anti-CD3 (BD Biosciences, San Jose, CA, USA), anti-B220 (BD Biosciences), anti-AA4.1 (eBioscience, San Diego, CA, USA), anti-IgM (eBioscience), anti-CD21 (BD Biosciences) and anti-CD23 (eBiosciences). The bone marrow cells were stained with anti-B220, anti-AA4.1 and anti-IgM. The cells were detected with a LSRII flow cytometer (BD Biosciences) or a FACScan (BD Biosciences) and analyzed with FlowJo software (Tree Star, Ashland, OR, USA).
CD23+ B-cell purification and stimulation
After treatment with red blood cell lysing buffer, spleen cells were incubated with anti-CD23-biotin (BD Biosciences) for 15min. The cells were washed with FACS buffer (3% fetal bovine serum, 2mM EDTA in PBS), incubated with streptavidin-magnetic particles (BD Biosciences) for 30min and then purified using iMagnet (BD Biosciences). The purified cells were analyzed by flow cytometry and were >95% B cells and 80% follicular B cells. Four hundred thousand cells were seeded in 96-well flat-bottom plates and stimulated with the relevant stimuli in complete RPMI 1640 (10% fetal bovine serum, 2mM l-glutamine, 50 µM 2-mercaptoethanol, 100U/ml penicillin and 100 µg/ml streptomycin) in the presence of 50ng/ml B lymphocyte stimulator (BLyS; R&D Systems, Minneapolis, MN, USA) in a total volume of 200 µl. After 48h, the supernatants were collected for cytokine measurement. For CD23+ B-cell proliferation, the cells were incubated with stimuli for 42h and then pulsed with 3H thymidine for 6h. Incorporation of 3H thymidine was quantified using a liquid scintillation beta counter.
DC preparation and stimulation
Bone marrow cells were seeded at 1.5 × 106 cells/ml in complete RPMI 1640 supplemented with 7.5% conditioned medium from B16 cells transfected with fml-like tyrosine kinase 3 ligand (Flt-3L). The Flt-3L B16 cells were originally made by Dr H. Chapman (36) and were provided by Dr U. Von Andrian (Harvard Medical School). The cells were used for experiments after 8 days, at which time >90% were CD11c-positive, of which 15–40% displayed a pDC phenotype (CD11c+ CD45RAhigh B220high CD11blow) and the remainder displayed a conventional DC (cDC) phenotype (CD11c+ CD45RAlow B220low CD11bhigh). Before setting up each assay, the Flt-3L DCs were routinely checked by flow cytometry for the relative percentages of pDCs and cDCs and the respective DC activation status after staining with anti-CD11c, anti-CD45RA, anti-B220, anti-CD11b and anti-CD62L (BD Biosciences). Flt-3L DCs (3 × 105 cells) were seeded in 96-well round-bottom plates and cultured in complete RPMI 1640 with the relevant stimuli in a total well volume of 200 µl. After 18–24h, the supernatants were collected for cytokine measurement.
Cytokine measurement
Cytokine levels in the cell culture supernatants were measured using in-house ELISAs developed using commercially available antibodies from BD Biosciences, PBL InterferonSource (Piscataway, NJ, USA) or US Biological (Swampscott, MA, USA) as previously described (6). The detection limit of the IL-6 ELISA is 16 pg/ml. The detection limit of the IFN-α ELISA is 80 pg/ml. The detection limit of the IFN-β ELISA is 160 pg/ml. IgG isotype and IgM levels in the sera were measured using in-house ELISAs developed using commercially available antibodies from BD Biosciences, Jackson ImmunoResearch and Southern Biotech (Birmingham, AL, USA). The detection limit for the IgG isotype and IgM ELISAs is 34 pg/ml.
Statistics
For in vitro experiments, groups were compared using a two-way Kruskal–Wallis analysis of variance on ranks and, in experiments where there were significant differences between groups (P < 0.05), the Holm–Sidak method was employed for post-hoc analysis and to correct for multiple comparisons. One-way analysis of variance on ranks was used to compare serum immunoglobulin levels of IRF5 +/+ mice to IRF5 −/− 11G mice with the DOCK2 mutation or IRF5 −/− 15G mice without the DOCK2 mutation, and the Dunn’s method was used for post-hoc analysis and to correct for multiple comparisons. All statistics were performed using SigmaPlot 11.0 (Systat, San Jose, USA) with P < 0.05 considered as statistically significant.
Results
IRF5−/− mice backcrossed 11 generations to C57BL/6 have a striking B-cell phenotype, which is normalized in IRF5−/− mice backcrossed further to C57BL/6
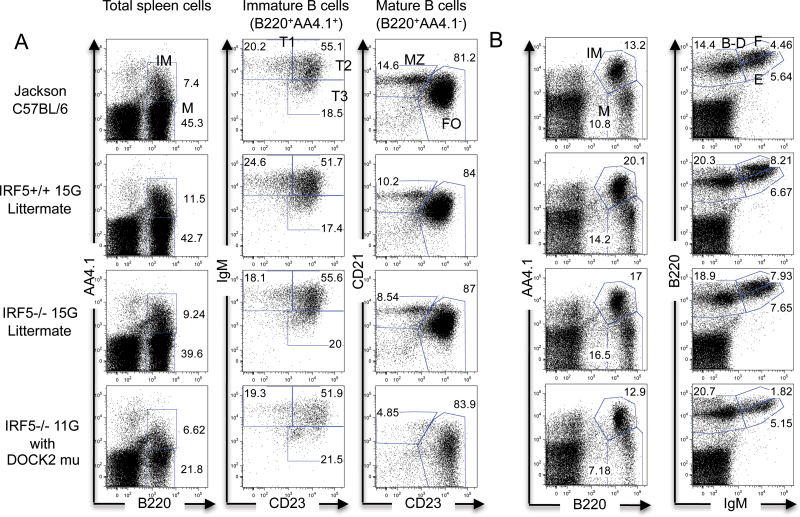
IRF family members are involved in lymphocyte development (1). To evaluate whether IRF5 plays a role in B-cell development, we initially used IRF5 −/− mice that had been backcrossed 11 generations to the C57BL/6 genetic background (hereafter termed IRF5 −/− 11G). Approximately 98.6% of the genome in these mice was C57BL/6 as determined using a panel of >500 000 SNPs (Table 2). We found that compared with C57BL/6 wild-type mice from Jackson Laboratory, IRF5 −/− 11G mice had a marked reduction in total splenic B-cell number with both mature and immature B-cell populations being reduced and an almost complete absence of splenic marginal zone B cells (Tables 3 and 4). In addition to a reduction in total B-cell number, there was also a reduction in the percentage of B cells relative to total (B and non-B) spleen cells. The number of splenic CD3+ T cells was also reduced although to a less marked extent (Table 3). IRF5 −/− 11G mice also exhibited abnormalities in bone marrow B cells with a reduction in Hardy fraction F (mature B cells or recirculating B cells) but not fractions B-D (pro-B and pre-B cells) or fraction E (immature B cells) (Fig. 1B).
Table 2.
Number and percentage of C57BL/6 and non-C57BL/6 SNPs in IRF5−/− mice
| SNPs | IRF5−/− 11G (with DOCK2 mutation) | IRF5−/− 14G (without DOCK2 mutation) |
|---|---|---|
| C57BL6 | 542 191 (98.64%) | 543 957 (98.96%) |
| Non-C57BL/6 | 1746 (0.32%) | 1315 (0.24%) |
| Heterozygous | 5677 (1.03%) | 4393 (0.8%) |
| Uncalled | 0 (0%) | 0 (0%) |
| Total | 549 614 (99.99%) | 549 665 (100%) |
Table 3.
Numbers and percentages of spleen cell populations
| Jackson C57BL/6 (n = 3) | IRF5+/+ 15G littermate (n = 4) | IRF5−/− 15G littermate (n = 3) | IRF5−/− 11G with DOCK2 mutation (n = 6) | |
|---|---|---|---|---|
| Total spleen cell number (×106) | 89.2±17.9 (100%) | 93.1±17.0 (100%) | 70.5±13.3 (100%) | 39.4±1.9 (100%) |
| B cells (B220+) | 47.9±10.3 (53.6%) | 48.1±10.4 (51.7%) | 34.1±7.2 (48.3%) | 10.4±0.9 (26.4%) |
| T cells (CD3+) | 29.2±6.3 (32.7%) | 30.0±2.9 (32.2%) | 25.2±4.0 (35.7%) | 17.4±1.0 (44.2%) |
| Non-B and non-T cells (B220–CD3–) | 7.7±0.7 (8.6%) | 9.1±2.1 (9.8%) | 6.4±0.7 (9.1%) | 9.3±0.3 (23.6%) |
Table 4.
Numbers and percentages of B-cell subpopulationsa in spleen
| Jackson C57BL/6 (n = 3) | IRF5+/+ 15G littermate (n = 4) | IRF5−/− 15G littermate (n = 3) | IRF5−/− 11G with DOCK2 mutation (n = 6) | |
|---|---|---|---|---|
| Total spleen cell number (×106) | 89.2±17.9 (100%) | 93.1±17.0 (100%) | 70.5±13.3 (100%) | 39.4±1.9 (100%) |
| Immature B cells | 8.0±1.5 (9.0%) | 9.5±1.4 (10.2%) | 6.4±0.8 (9.1%) | 2.9±0.3 (7.3%) |
| T1 | 1.7±0.4 (1.9%) | 2.1±0.2 (2.3%) | 1.1±0.3 (1.6%) | 0.4±0.0 (1%) |
| T2 | 4.2±0.8 (4.7%) | 5.1±0.7 (5.5%) | 3.6±0.6 (5.1%) | 1.4±0.1 (3.6%) |
| T3 | 1.4±0.4 (1.6%) | 1.6±0.5 (1.7%) | 1.2±0.3 (1.7%) | 0.7±0.2 (1.8%) |
| Mature B cells | 42.7±9.5 (47.9%) | 43.5±10.9 (46.7%) | 32.2±8.4 (45.7%) | 9.7±0.9 (24.6%) |
| Follicular B cells | 37.1±9.4 (41.6%) | 38.3±10.3 (41.1%) | 28.8±7.7 (40.9%) | 8.6±0.9 (21.8%) |
| Marginal zone B cells | 4.0±0.2 (4.4%) | 3.2±0.4 (3.4%) | 2.3±0.5 (3.3%) | 0.3±0.0 (0.8%) |
aThe B-cell subpopulations were identified as shown in Fig. 1.
Fig. 1.
Normal percentages of B-cell populations in the spleen and bone marrow of IRF5 −/− 15G mice. (A) Spleen cells from Jackson C57BL/6 wild-type mice, IRF5 +/+ and IRF5 −/− 15G littermates and IRF5 −/− 11G mice (with the DOCK2 mutation) were stained with antibodies against B220, AA4.1, IgM, CD23 and CD21. Percentages of mature (M) B cells (B220+ AA4.1-) and immature (IM) B cells (B220+ AA4.1+) were determined. Immature B cells were further classified as transitional 1 (T1), T2 or T3 based on IgM and CD23 expression. Mature B cells were further classified as marginal zone (MZ) or follicular (FO) B cells based on CD21 and CD23 expression. The data are representative of three to six independent experiments. (B) Bone marrow cells from the same mice evaluated in (A) were stained with antibodies against B220, AA4.1 and IgM to detect mature B cells (B220+ AA4.1-) and immature, pro-B and pre-B B cells (B220+ AA4.1+) (left-hand panels). The relative percentages of Hardy fractions B-D (pro-B and pre-B; B220+ IgM-), fraction E (immature B; B220intermediate IgM+) and fraction F (mature B; B220high IgM+) were also determined (right-hand panels) (37). The data are representative of three independent experiments.
We initially assumed that these differences were due to the deficiency of IRF5, and thus, we were surprised to find that the observed B-cell phenotype was much less apparent when the IRF5 −/− 11G mice were backcrossed a further three or four generations to C57BL/6 (hereafter referred to as IRF5 −/− 14G and IRF5 −/− 15G, respectively). We, therefore, concluded that there must be another gene(s) that had contributed to the B-cell phenotype but that had been lost during the further backcrossing. To determine the role of IRF5 in B-cell development in the absence of this confounding gene, we repeated the analysis using IRF5 +/+ and IRF5 −/− 15G littermates derived from matings between IRF5 +/- 15G heterozygous mice. There was no difference between IRF5 +/+ 15G and IRF5 −/− 15G littermates in the percentages of total splenic B cells or the various B-cell populations including the marginal zone B cells (Fig. 1A and Tables 3 and 4). There was also no difference between IRF5 +/+ 15G and IRF5 −/− 15G littermates in the percentages of the various Hardy fractions in the bone marrow (Fig. 1B). Overall, these findings suggested that the observed decrease in B-cell number and loss of marginal zone B cells in IRF5 −/− 11G mice was not due to IRF5 deficiency.
IRF5−/− 11G mice have a mutation in DOCK2, whereas IRF5−/− 14G and IRF5−/− 15G mice do not
At the time we were doing these studies, Drs Diamond and Bhattacharya informed us that they had found a similar B-cell phenotype in their IRF5 −/− mice to that which we had observed and that an exon 28–29 duplication in the DOCK2 gene in the IRF5 −/− mice was the cause of this phenotype (31). Based on our B-cell phenotype data, it was therefore likely that the IRF5 −/− 11G mice would have this mutation in DOCK2, whereas the IRF5 −/− 14G and IRF5 −/− 15G mice would not, and we confirmed that this was indeed the case using an RT–PCR assay designed by Drs Diamond and Bhattacharya (31) (Fig. 2A).
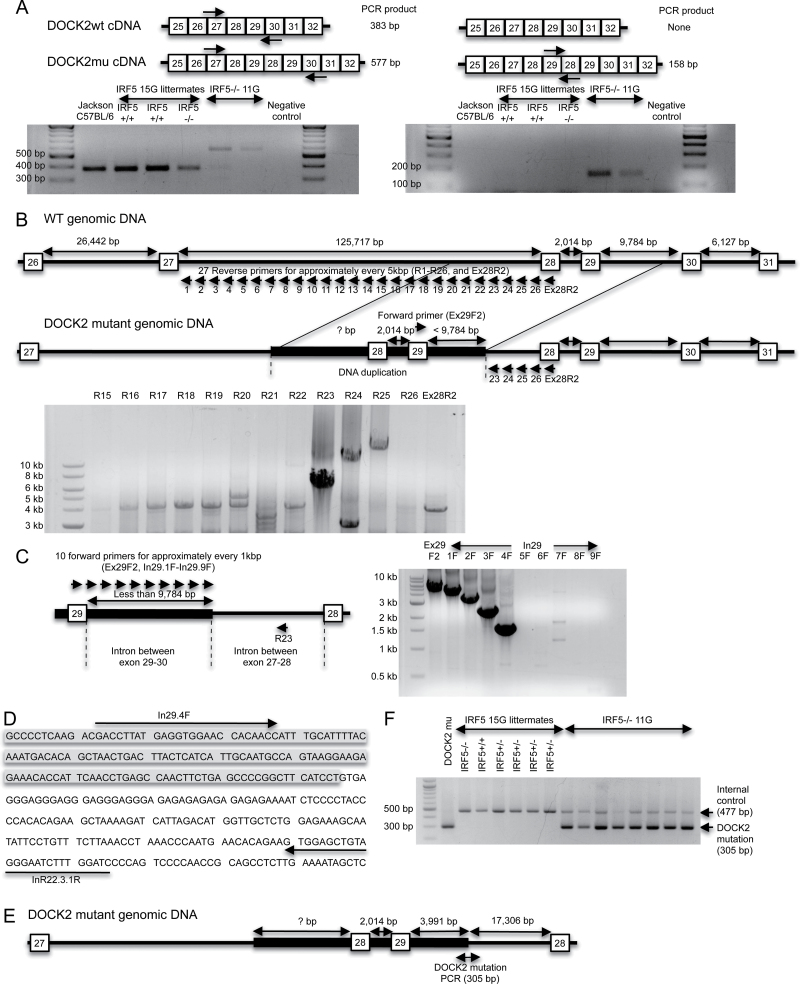
Fig. 2.
A DOCK2 mutation is detected in IRF5 −/− mice backcrossed 11 generations to C57BL/6 (IRF5 −/− 11G) mice but not in IRF5 −/− mice backcrossed 15 generations to C57BL/6 (IRF5 −/− 15G). (A) RT–PCR to detect the DOCK2 mutation (DOCK2mu). RNA was purified from splenic B cells of wild-type C57BL/6 mice from The Jackson Laboratory (Jackson wild-type), IRF5 +/+ and IRF5 −/− 15G littermates and IRF5 −/− 11G mice. RNA was reverse transcribed into cDNA and amplified using specific primers to detect the exon 28–29 duplication as described (31). Primers used in the left-hand gel give a 577-bp product for the DOCK2 mutation and a 383-bp product for wild-type DOCK2. Primers used in the right-hand gel give a 158-bp product for the DOCK2 mutation and do not amplify wild-type DOCK2. (B) The diagram shows the hypothesized differences in genomic DOCK2 DNA between wild-type and DOCK2 mutant IRF5 −/− mice. In the DOCK2 mutant mice, the duplicated exon 28 and 29 together with some flanking DNA is inserted into the intron between exons 27 and 28. The gel shows a PCR performed using a forward primer, which recognizes exon 29 (Ex29F2) and 27 reverse primers (R1-R26 or Ex28R2), which detect the region in the intron between exons 27 and 28 that is closest to exon 28. PCR products were obtained with the R23–R25 primers. IRF5 −/− 11G genomic DNA containing the DOCK2 mutation was used as the template. (C) PCR was performed using the R23 reverse primer and 10 forward primers, which recognize either exon 29 (Ex29F2) or the intron between exons 29 and 30 (In\.1F to In29.9F). (D) DNA sequence of the 3′-end of DOCK2 mutation. The shaded region is the duplicated intronic sequence between exons 29 and 30, and the unshaded region is the non-duplicated intron between exons 27 and 28. (E) Diagram of the DOCK2 mutation. The duplicated segment of the DOCK2 gene present in the DOCK2 mutation ends at 3991bp after exon 29. This duplicated segment is inserted into intron 27–28 at 17 306bp before exon 28. (F) PCR to detect the DOCK2 mutation. Genomic DNA from IRF5 −/− 11G mice gave a PCR product for the DOCK2 mutation (305bp), whereas DNA from IRF5 +/+ and IRF5 −/− 15G littermates did not. CD19 PCR was used as an internal control to verify the adequacy of DNA preparation in each sample.
We next wanted to determine the extent to which the DOCK2 mutation was present in our IRF5 knockout line. RT–PCR is not an ideal screening assay as it requires mRNA isolation from hematopoietic cells and is more labor intensive than a DNA-based screen. Furthermore, RT–PCR cannot be used to determine the DOCK2 genotype of mice used in previous experiments unless RNA from hematopoietic cells happened to be saved from that particular experiment. Therefore, it was important to develop an efficient and reproducible PCR assay to detect the DOCK2 mutation in genomic DNA. To do this, it was first necessary to determine the insertion end-point of the DOCK2 mutation within genomic DNA.
The mutation in the DOCK2 gene is an exon 28–29 duplication that results in a substantial reduction in DOCK2 mRNA expression (31). We hypothesized that the duplicated DNA might be inserted somewhere before the original genomic exon 28 and must include some segment of the intron sequence between exon 27 and 28, as well as some segment of the intron sequence between exon 29 and 30 (Fig. 2B). Because the length of the intron sequence between exon 29 and 30 in the wild-type DOCK2 gene is 9784bp, the distance between the duplicated exon 29 and the end-point of the insertion must be <9784bp. We designed a forward PCR primer in exon 29 (termed Ex29F2) and 26 reverse PCR primers for every 5kb in the 125 717bp intron between exons 27 and 28 (termed R1–26) and one reverse PCR primer on exon 28 (termed Ex28R2). Using those primers, we performed long-range PCR using DNA polymerase that can amplify up to 30kb (Fig. 2B). DNA electrophoresis showed that three reverse PCR primers (R23, R24 and R25) amplified DNA, whereas primers R1–22, R26 and Ex28R2 did not, suggesting that the 3′ end-point of the inserted DOCK2 duplication is between the R22 and R23 primer recognition sites (Fig. 2B). Using the R23 primer, we next sought to determine the length of the duplicated intronic segment between the duplicated exon 29 and this 3′ end-point. We designed nine additional forward primers for every 1kb after the duplicated exon 29 (termed In29.1F to 9F) and did long-range PCR using R23 as a reverse primer (Fig. 2C). Primers from Ex29F2 and In29.1F to In29.4F successfully amplified DNA from genomic DNA of mice with the DOCK2 mutation, whereas primers In29.5F to In29.9F did not (Fig. 2C). This result suggested that the duplicated DOCK2 insertion ends somewhere between the recognition site of In29.4F and In29.5F. DNA sequencing of the PCR product demonstrated that the DOCK2 duplication ended 3991bp after the duplicated exon 29, and this occurred at a position 17 306bp before the non-duplicated exon 28 (Fig. 2D and E).
On the basis of this information, we designed a final PCR protocol that detects the DOCK2 mutation in genomic DNA and gives a PCR product of 305bp (Fig. 2F). This PCR demonstrated that the IRF5 −/− 11G mice had the mutation in DOCK2, whereas the IRF5 −/− 14G and IRF5 −/− 15G mice did not (Fig. 2F and data not shown) confirming the previous results obtained using RT–PCR (Fig. 2A).
IRF5 is involved in B-cell IL-6 production in response to TLR7 and TLR9 ligands, whereas the DOCK2 mutation is involved in B-cell IL-6 production in response to TLR2 and TLR4 ligands
To determine the relative contributions of IRF5 and the DOCK2 mutation to B-cell functional responses, we next evaluated B-cell cytokine production and proliferation. Cytokine production in response to TLR stimulation was compared using B cells from wild-type C57BL/6, IRF5 −/− 11G and IRF5 −/− 14G mice. Because IRF5 −/− 11G mice have no marginal zone B cells in the spleen but IRF5−/− 14G mice do, we purified B cells using anti-CD23 antibody positive selection, which excludes marginal zone B cells but includes other splenic B-cell populations. Purified B cells from all groups of mice had equal percentages of immature B cells (B220+ AA4.1+), mature B cells (B220+ AA4.1-) and follicular B cells (B220+ AA4.1- CD23+) but no marginal zone B cells (B220+ AA4.1- CD21+ CD23-) (Fig. 3A).
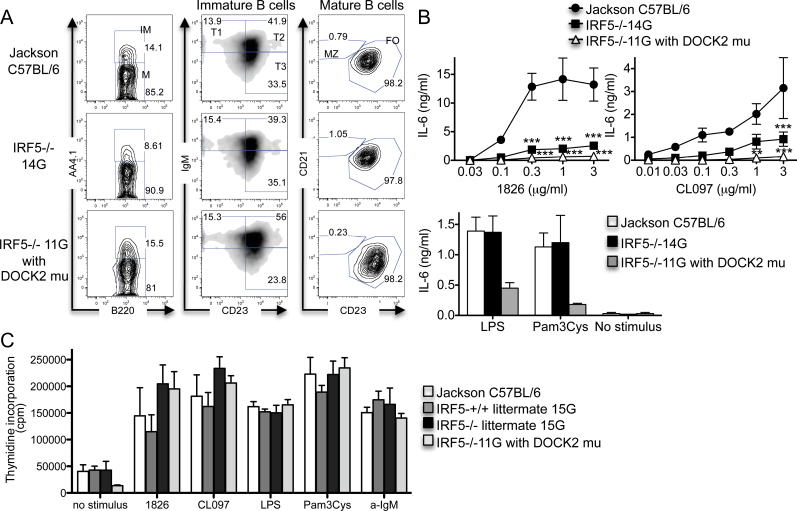
Fig. 3.
Relative contributions of IRF5 and the DOCK2 mutation to B-cell IL-6 production and proliferation. (A and B) B cells were purified from the spleens of Jackson C57BL/6 wild-type mice, IRF5 −/− 14G mice and IRF5 −/− 11G mice (with the DOCK2 mutation) using CD23-positive selection. (A) The B cells were stained with antibodies against B220, AA4.1, IgM, CD21 and CD23. Data are representative of three independent experiments. IM, immature B cells; M, mature B cells; T, transitional B cells; MZ, marginal zone B cells; FO, follicular B cells. (B) B cells were stimulated with LPS (30 µg/ml), Pam3Cys (1 µg/ml), 1826 and CL097 for 48h in the presence of BLyS (50ng/ml). IL-6 levels in the supernatants were measured by ELISA. The data represent the mean ± SEM of three independent experiments. **P < 0.01; ***P < 0.001 versus IRF5+/+. There was no statistical difference between B cells from IRF5 −/− 14G and IRF5 −/− 11G mice. (C) B cells were purified from the spleens of Jackson C57BL/6 wild-type mice (n = 3), IRF5 +/+ (n = 5) and IRF5 −/− (n = 3) 15G littermates, and IRF5 −/− 11G mice (with the DOCK2 mutation) (n =6) using CD23-positive selection. The B cells were stimulated with 1826 (3 µg/ml), CL097 (3 µg/ml), LPS (30 µg/ml), Pam3Cys (1 µg/ml) and anti-IgM antibody (30 µg/ml) for 42h in the presence of BLyS (50ng/ml) and pulsed with 3H thymidine for 6h. The data represent the mean ± SEM of three independent experiments.
We stimulated the anti-CD23-purified B cells with TLR ligands for 48h and measured IL-6 production. B cells from both IRF5 −/− 11G (with the DOCK2 mutation) and IRF5 −/− 14G (without the DOCK2 mutation) mice produced less IL-6 than wild-type mice in response to the TLR7 ligand CL097 and the TLR9 ligand CpG-B. In contrast, and surprisingly, IL-6 production in response to the TLR2 ligand Pam3Cys and the TLR4 ligand LPS was reduced only in B cells from IRF5 −/−11G mice (Fig. 3B). These results indicate that IRF5 is involved in IL-6 production downstream of TLR7 and TLR9 signaling in CD23+ B cells, but that the DOCK2 mutation, and not IRF5, limits IL-6 production downstream of TLR2 and TLR4 signaling in CD23+ B cells.
To determine whether IRF5 or the DOCK2 mutation played a role in B-cell proliferation, we compared responses of anti-CD23-purified B cells from IRF5 +/+ and IRF5 −/− 15G littermates and IRF5 −/− 11G mice. Proliferative responses induced by anti-IgM and TLR ligands were similar in B cells from IRF5 −/− 11G mice and IRF5 −/− 15G mice indicating that the DOCK2 mutation does not contribute to these responses, although baseline proliferation in the absence of any stimulus was lower in B cells from the IRF5 −/− 11G mice (Fig. 3C). Unexpectedly, B cells from IRF5 −/− 15G mice proliferated slightly more strongly to TLR7 and TLR9 ligands than B cells from their IRF5 +/+ 15G littermates, suggesting the possibility that IRF5 may act to inhibit B-cell proliferation downstream of TLR7 and TLR9. This was not due to a generalized reduction of proliferation in the IRF5 +/+ B cells as there was no difference between B cells from IRF5 +/+ and IRF5 −/− 15G mice in proliferative responses to anti-IgM or LPS or in basal proliferation in the absence of stimulus (Fig. 3C).
IRF5 is involved in IFN-α , IFN−β and IL-6 production by murine DCs downstream of TLR9 and/or TLR7
Mouse bone marrow cells cultured in vitro with Flt3 ligand develop into a mixed population of pDCs and cDCs, collectively referred to as Flt-3L DCs (38). Previously, we have shown that IRF5 is involved in the production of IFN-α, IFN-β and IL-6 from mouse Flt-3L DCs downstream of TLR7 and TLR9, but not downstream of TLR2 and TLR4 (6). As the IRF5 −/− mice used in these previous studies likely had the DOCK2 mutation and pDCs from DOCK2-deficient mice have a reduced capacity to make IFN-α and IFN-β after TLR9 and TLR7 activation (35), it was important to determine whether our previously reported results were due to IRF5 deficiency or to the DOCK2 mutation. To do this, we prepared Flt-3L DCs from IRF5 +/+ and IRF5 −/− 15G littermates and IRF5 −/− 11G mice using an identical procedure to that used previously. There was no difference between the three experimental groups in the percentages of cDCs and pDCs in the Flt-3L DC cultures (Fig. 4A).
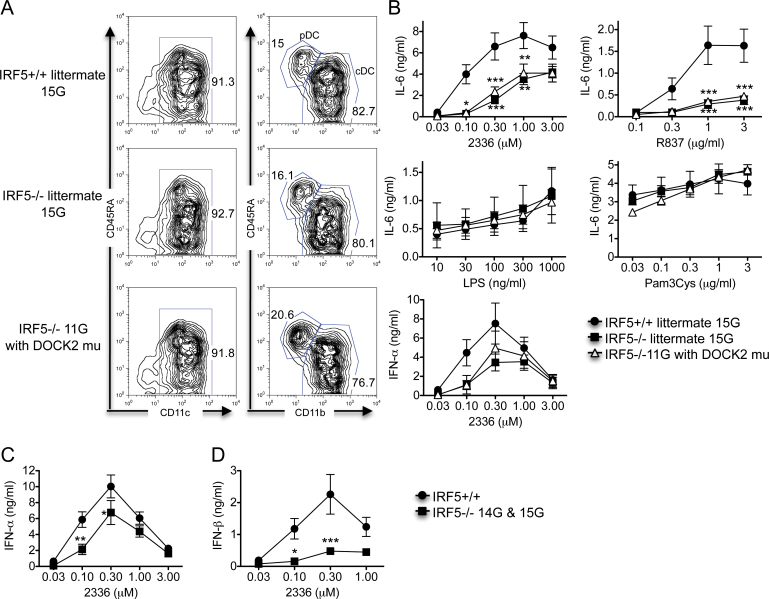
Fig. 4.
IRF5 is required for TLR7- and TLR9-induced IL-6 and type I interferon production by Flt-3L DCs. (A and B) Bone marrow-derived Flt-3L DCs were prepared from IRF5 +/+ (n = 5) and IRF5 −/− 15G littermates (n = 3) and IRF5 −/− 11G mice (with the DOCK2 mutation) (n = 6). (A) Flt-3L DCs were stained with antibodies to CD11c, CD45RA and CD11b to determine the relative percentages of pDCs (CD11c+, CD45RAhigh and CD11b-) and cDCs (CD11c+, CD45RAlow and CD11b+). The data are representative of three independent experiments. (B) Flt-3L DCs were stimulated with the TLR9 ligand CpG-A (2336), the TLR7 ligand R837, the TLR4 ligand LPS and the TLR2 ligand Pam3Cys for 24h. The levels of IFN-α and IL-6 in the supernatants were measured by ELISA. The data represent the mean ± SEM of three independent experiments. (C and D) Bone marrow-derived Flt-3L DCs were prepared from IRF5+/+ (n = 10; either IRF5+/+ 15G littermates or Jackson C57BL/6) and IRF5−/− (n = 7; either IRF5−/− 15G littermates or IRF5−/− 14G mice). Flt-3L DCs were stimulated with the TLR9 ligand CpG-A (2336) for 24h. The levels of IFN-α (C) and IFN-β (D) in the supernatants were measured by ELISA. The data represent the mean ± SEM of 6–7 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 versus IRF5+/+.
Flt-3L DCs from both IRF5 −/− 15G and IRF5 −/− 11G mice produced less IL-6 than Flt-3L DCs from IRF5 +/+ mice after activation by TLR9 and TLR7 ligands with no difference seen in IL-6 production between the IRF5 −/− 15G and IRF5 −/− 11G groups (Fig. 4B). These results indicate that IRF5 is involved in TLR9- and TLR7-induced IL-6 production by Flt-3L DCs. No differences were seen between any of the three experimental groups in IL-6 production induced by TLR2 and TLR4 ligands.
Flt-3L DCs from both IRF5 −/− 15G and IRF5 −/− 11G mice produced less IFN-α than Flt-3L DCs from IRF5 +/+ mice after stimulation with CpG-A concentrations of 0.1 and 0.3 µM but not after stimulation with higher concentrations of CpG-A. There was no difference in IFN-α production between IRF5 −/− 15G and IRF5 −/− 11G mice (Fig. 4B). To confirm these findings, we performed additional experiments comparing Flt-3L DCs from Jackson C57BL/6 wild-type (IRF5 +/+) mice with Flt-3L DCs from IRF5 −/− 14G or 15G mice and combined these data with the IRF5 +/+ and IRF5 −/− 15G littermate data. The combined data mirrored the IRF5 +/+ and IRF5 −/− 15G littermate data and demonstrated reduced IFN-α production by the IRF5 −/− Flt-3L DCs at CpG-A concentrations of 0.1 and 0.3 µM (Fig. 4C). IFN-β production by the IRF5 −/− Flt-3L DCs was even more dramatically reduced (Fig. 4D). Overall, these results are very similar to those we previously reported (6) and demonstrate that IRF5 is involved in TLR9-induced IFN-α and IFN-β production by Flt-3L DCs.
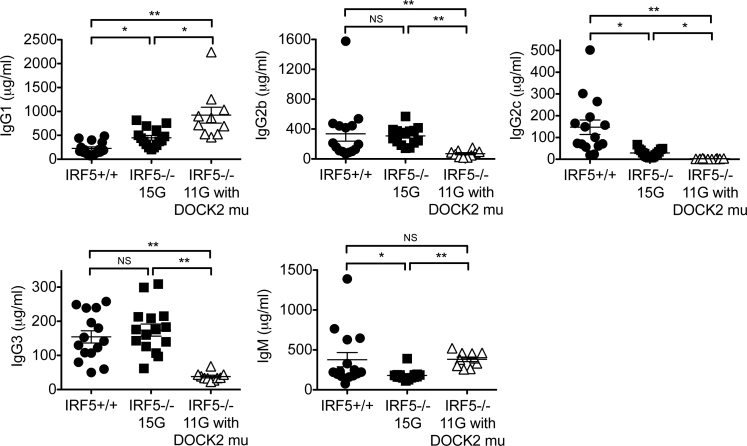
The presence of the DOCK2 mutation enhances Th2 skewed IgG production in IRF5−/− mice
Several reports using IRF5 −/− mice have shown that IRF5 is required for a normal Th1 response, while IRF5 −/− mice show a more skewed Th2 response (30, 39, 40). However, it has also been shown that DOCK2-deficient mice have an enhanced Th2-type response and a suppressed Th1 response (34). The authors observed that DOCK2 −/− mice have higher serum levels of IgG1 and IgE, but a lower level of IgG2b. Therefore, it is critical to determine whether the Th1/Th2 imbalance observed in IRF5 −/− mice with the DOCK2 mutation is a result of IRF5 deficiency or the presence of the DOCK2 mutation. To examine this, we measured serum IgG isotype and IgM levels in wild-type mice, IRF5 −/− 11G mice with the DOCK2 mutation and IRF5 −/− 15G mice without the DOCK2 mutation. IRF5 −/− 15G mice had less IgG2a/c and IgM and slightly higher levels of IgG1 compared with IRF5 +/+ mice, but there was no difference in IgG2b and IgG3 levels (Fig. 5) This suggests that, as previously reported, IRF5 is involved in class switching to the IgG2a/c isotype (39). However, IRF5 −/− 11G mice with the DOCK2 mutation have dramatically increased levels of IgG1 compared with IRF5 +/+ mice and IRF5 −/− 15G mice (Fig. 5). In addition, IRF5 −/− 11G mice with the DOCK2 mutation have less IgG2b, IgG2a/c and IgG3 compared with IRF5 −/− 15G mice without the DOCK2 mutation. These results clearly show that Th2 skewing of IgG isotypes is enhanced in IRF5-deficient mice by the presence of the DOCK2 mutation.
Fig. 5.
Serum IgG isotype and IgM levels. IgG isotype and IgM levels in the sera of IRF5 +/+ 15G mice (n = 15), IRF5 −/− 15G mice (n = 15) and IRF5 −/− 11G mice with the DOCK2 mutation (n = 10) were measured by ELISA. *P < 0.05; **P< 0.01.
Discussion
IRF5 −/− mice have been used for many studies of IRF5 biology (5–9, 27–29, 39–46). Purtha et al. (31) have recently shown that there is a DOCK2 mutation present in a commonly distributed IRF5 −/− line that results in a reduced level of DOCK2 expression. It is, therefore, important to be able to easily identify IRF5 −/− mice that have, and do not have, the DOCK2 mutation so that archived tissue from past experiments can be regenotyped and future experiments can be done using IRF5 −/− mice without the mutation. Where archived tissue is not available, it will also be necessary to repeat certain studies to determine whether effects attributed to IRF5 in previously published reports were in fact due to IRF5 rather than to the DOCK2 mutation.
Purtha et al. (31) used an RT–PCR assay to detect the DOCK2 mutation. This assay was invaluable in the initial characterization of the DOCK2 mutation, but it does have certain limitations. Firstly, it requires RNA that needs to be obtained from DOCK2-expressing hematopoietic cells. Although RNA can be obtained from peripheral blood, this is not ideal for routine screening in living mice. Secondly, RNA is often not available from experiments that have been performed in the past. Thus, if investigators want to determine whether the DOCK2 mutation was present in the IRF5 −/− mice used for these previous experiments, it may not be possible to do this using RT–PCR. In order to overcome these limitations, we determined the insertion point of the exon 28–29 duplication and designed a PCR assay to detect the DOCK2 mutation in genomic DNA. This PCR can be used to detect the DOCK2 mutation in DNA obtained from tail samples or other tissue samples. One limitation of our assay is that it cannot be used to determine whether the mutation is heterozygous or homozygous. Nevertheless, we have been able to use the PCR to identify IRF5 −/− mice that do not have the mutation and thus obtain a colony of IRF5 −/− mice without the DOCK2 mutation.
IRF5 −/− 11G mice (that have the DOCK2 mutation) exhibited a marked reduction in total spleen cell number. This was due mainly to a reduction in both the percentage and number of splenic B cells although some reduction in T-cell number was also evident. IRF5 −/− 15G mice (that do not have the DOCK2 mutation) exhibited a normal percentage of splenic B cells and a substantial increase in splenic B-cell number compared with IRF5 −/− 11G mice suggesting that the DOCK2 mutation was responsible for these observed differences between IRF5 −/− 11G and IRF5 −/− 15G mice. This interpretation is consistent with the findings of Purtha et al. (31) as well as with the known role of DOCK2 in trafficking of B cells, T cells and neutrophils (32, 33).
Previous studies using B cells from IRF5 −/− mice have shown that IL-6 production induced by TLR9 and TLR7 ligands is IRF5 dependent (5, 43). We have confirmed these findings using B cells from IRF5 −/− 14G mice that do not express the DOCK2 mutation. We also found that, in contrast to its role in TLR9- and TLR7-induced IL-6 production, IRF5 is not involved in IL-6 production downstream of TLR2 or TLR4 in B cells. Unexpectedly however, IL-6 production downstream of TLR2 or TLR4 was much lower in B cells from IRF5 −/− 11G mice, but not IRF5 −/−15G mice, suggesting the possibility that DOCK2 might be involved in TLR2- and TLR4-induced B-cell cytokine production. To the best of our knowledge, the role of DOCK2 in TLR-induced cytokine production in B cells has not been studied previously but could be examined in future studies using B cells from DOCK2-knockout mice.
Using IRF5 +/+ and IRF5 −/− 15G littermates, we found that TLR9 (CpG-A)-induced IFN-α production by pDCs within Flt-3L DC cultures is IRF5 dependent at lower CpG-A concentrations consistent with our previous observation (6). Importantly, IFN-β production induced by CpG-A was markedly reduced in IRF5 −/− Flt-3L DCs compared with IRF5 +/+ Flt-3L DCs, emphasizing the underappreciated role of IRF5 in IFN-β production. Overall, our data demonstrate that IRF5 plays a role in TLR9-induced type I interferon production independent of the DOCK2 mutation. This conclusion is different from that reached by Purtha et al. (31) who concluded that type I interferon responses are largely intact in IRF5 −/− DCs. This may reflect the fact that IFN-β production was not evaluated in that report. Given the pleiotropic immunomodulatory effects of type I interferons (47), induction of IFN-α and IFN-β by IRF5 may represent one important way whereby IRF5 regulates immune responses during infection as well as in autoimmune disease.
We found that IRF5 deficiency alone leads to a slight increase in serum IgG1 production and a large decrease in IgG2a/c production in C57BL/6 mice although IRF5 deficiency had no effect on the levels of the other Th1-induced isotypes IgG2b and IgG3. This finding is consistent with the reported ability of IRF5 to specifically bind to the IgG2a/c promoter in B cells (39) and suggests that IRF5 deficiency may induce a mild Th2-like phenotype, at least in the absence of infection or autoimmunity. However, we found that in IRF5 −/− 11G mice with the DOCK2 mutation, there was a dramatic skewing toward Th2-type IgG isotypes with much more IgG1 production and much less IgG2b, IgG2a/c and IgG3 production than in their IRF5 −/− DOCK2 wild-type counterparts. This is consistent with a previous report showing that DOCK2 deficiency results in skewing toward a Th2 response because CD4+ T cells in DOCK2 −/− mice cannot down-regulate IL-4Rα surface expression due to the lack of DOCK2-dependent Rac activation in CD4+ T cells (34). Overall, the DOCK2 mutation greatly enhances the mild Th2 phenotype of IRF5 −/− mice.
We previously reported that heterozygous and homozygous deficiency of IRF5 markedly reduced disease severity in the FcγRIIB −/− Yaa male and FcγRIIB −/− female mouse models of systemic lupus erythematosus (27). These mice were generated by intercrossing IRF5-deficient mice backcrossed eight generations to C57BL/6 with FcγRIIB −/− Yaa mice. To determine the extent to which the DOCK2 mutation was present in these original experimental mice, we purified DNA from archived tail samples and analyzed the DNA using the PCR described in Fig. 2. All samples from the FcγRIIB −/− Yaa male experimental cohorts (n = 33) were positive for the DOCK2 mutation, except for one. In the FcγRIIB −/− female experimental cohorts, a total of six samples tested negative for the DOCK2 mutation: four out of 15 FcγRIIB −/− IRF5 +/+, one out of 21 FcγRIIB −/− RII IRF5 +/− and one out of 22 FcγRIIB −/− IRF5 −/−. A limited analysis suggested that IRF5 deficiency, and not the DOCK2 mutation, was likely responsible for the protective phenotype observed in the FcγRIIB −/− IRF5 +/− and FcγRIIB −/− IRF5 −/− mice (data not shown). However, given the very small number of mice without the DOCK2 mutation, it will be important to confirm the protective effect of IRF5 deficiency in the FcγRIIB −/− model by evaluating new cohorts of FcγRIIB −/− IRF5 +/+, IRF5 +/− and IRF5 −/− mice that do not have the DOCK2 mutation.
The origin of the DOCK2 mutation is not known. However, the fact that the DOCK2 mutation was observed in two separate IRF5 −/− colonies at Boston University and Johns Hopkins University initiated from mice obtained independently from the original colony suggests that at least a subset of IRF5 −/− mice from the original colony possessed the DOCK2 mutation (31). It is possible that the DOCK2 mutation arose as a spontaneous mutation in the IRF5 −/− line. Alternatively, it is possible that the DOCK 2 mutation was present in C57BL/6 mice used in the backcrossing of the IRF5 −/− mice or was present in the embryonic stem cells used in the generation of the IRF5 −/− mice. If the latter is the case, the DOCK2 mutation may not be restricted to the IRF5 −/− line and may be more widespread in other experimental mice. There is a precedent for this possibility. For example, caspase 1 knockout mice and cellular inhibitor of apoptosis 1 knockout mice are also deficient in caspase 11 expression as a result of a mutation in the caspase 11 locus in the 129 embryonic stem cells used to make these mice (48, 49). It is also not clear why the DOCK2 mutation has persisted in the IRF5 −/− line despite backcrossing to the C57BL/6 background given that the IRF5 locus and the DOCK2 locus are unlinked.
In future studies, it will be necessary to more completely define the biological functions of IRF5 both in vitro and in vivo. The availability of a fully backcrossed IRF5 −/− line without the DOCK2 mutation and a PCR assay able to reliably detect the DOCK2 mutation in genomic DNA should facilitate these efforts.
Funding
National Institutes of Health (P01 AR050256 to I.R.R. and A.M-R., 1K01AR060857-01 to K.Y., and R01 DK090558 to R.G.B.); Research Training in Nephrology T32 Grant from National Institutes of Health (T32DK007053 to K.Y.) and Research Training in Immunology T32 Grant from National Institutes of Health (AI007309-23 to A.A.W.).
Acknowledgements
We thank Drs Diamond and Bhattacharya for sharing their findings on DOCK2 mutation with us prior to publication. We also thank Gabriella E. Wilson for technical assistance.
References
- 1. Tamura T., Yanai H., Savitsky D., Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26: 535 [DOI] [PubMed] [Google Scholar]
- 2. Paun A., Pitha P. M. 2007. The IRF family, revisited. Biochimie 89: 744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes B. J., Moore P. A., Pitha P. M. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276: 23382 [DOI] [PubMed] [Google Scholar]
- 4. Barnes B. J., Kellum M. J., Field A. E., Pitha P. M. 2002. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol. Cell. Biol. 22: 5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takaoka A., Yanai H., Kondo S., et al. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434: 243 [DOI] [PubMed] [Google Scholar]
- 6. Yasuda K., Richez C., Maciaszek J. W., et al. 2007. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J. Immunol. 178: 6876 [DOI] [PubMed] [Google Scholar]
- 7. Yanai H., Chen H. M., Inuzuka T., et al. 2007. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 104: 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pandey A. K., Yang Y., Jiang Z., et al. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis . PLoS Pathog. 5: e1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Couzinet A., Tamura K., Chen H. M., et al. 2008. A cell-type-specific requirement for IFN regulatory factor 5 (IRF5) in Fas-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105: 2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu G., Barnes B. J. 2009. IRF-5 is a mediator of the death receptor-induced apoptotic signaling pathway. J. Biol. Chem. 284: 2767 [DOI] [PubMed] [Google Scholar]
- 11. Sigurdsson S., Nordmark G., Göring H. H., et al. 2005. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am. J. Hum. Genet. 76: 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graham R. R., Kozyrev S. V., Baechler E. C., et al. Argentine and Spanish Collaborative Groups 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 38: 550 [DOI] [PubMed] [Google Scholar]
- 13. Dieudé P., Guedj M., Wipff J., et al. 2009. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 60: 225 [DOI] [PubMed] [Google Scholar]
- 14. Radstake T. R., Gorlova O., Rueda B., et al. Spanish Scleroderma Group 2010. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 42: 426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miceli-Richard C., Comets E., Loiseau P., Puechal X., Hachulla E., Mariette X. 2007. Association of an IRF5 gene functional polymorphism with Sjögren’s syndrome. Arthritis Rheum. 56: 3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson C. A., Boucher G., Lees C. W., et al. 2011. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43: 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dieguez-Gonzalez R., Calaza M., Perez-Pampin E., et al. 2008. Association of interferon regulatory factor 5 haplotypes, similar to that found in systemic lupus erythematosus, in a large subgroup of patients with rheumatoid arthritis. Arthritis Rheum. 58: 1264 [DOI] [PubMed] [Google Scholar]
- 18. Stahl E. A., Raychaudhuri S., Remmers E. F., et al. BIRAC Consortium; YEAR Consortium 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42: 508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cunninghame Graham D. S., Manku H., Wagner S., et al. 2007. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum. Mol. Genet. 16: 579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham R. R., Kyogoku C., Sigurdsson S., et al. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. U.S.A. 104: 6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferreiro-Neira I., Calaza M., Alonso-Perez E., et al. 2007. Opposed independent effects and epistasis in the complex association of IRF5 to SLE. Genes Immun. 8: 429 [DOI] [PubMed] [Google Scholar]
- 22. Marshak-Rothstein A., Rifkin I. R. 2007. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 25: 419 [DOI] [PubMed] [Google Scholar]
- 23. Christensen S. R., Shupe J., Nickerson K., Kashgarian M., Flavell R. A., Shlomchik M. J. 2006. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25: 417 [DOI] [PubMed] [Google Scholar]
- 24. Pisitkun P., Deane J. A., Difilippantonio M. J., Tarasenko T., Satterthwaite A. B., Bolland S. 2006. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312: 1669 [DOI] [PubMed] [Google Scholar]
- 25. Deane J. A., Pisitkun P., Barrett R. S., et al. 2007. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27: 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Z., Davidson A. 2012. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat. Med. 18: 871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richez C., Yasuda K., Bonegio R. G., et al. 2010. IFN regulatory factor 5 is required for disease development in the FcgammaRIIB-/-Yaa and FcgammaRIIB-/- mouse models of systemic lupus erythematosus. J. Immunol. 184: 796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tada Y., Kondo S., Aoki S., et al. 2011. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 63: 738 [DOI] [PubMed] [Google Scholar]
- 29. Xu Y., Lee P. Y., Li Y., et al. 2012. Pleiotropic IFN-dependent and -independent effects of IRF5 on the pathogenesis of experimental lupus. J. Immunol. 188: 4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng D., Yang L., Bi X., Stone R. C., Patel P., Barnes B. J. 2012. Irf5-deficient mice are protected from pristane-induced lupus via increased Th2 cytokines and altered IgG class switching. Eur. J. Immunol. 42: 1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Purtha W. E., Swiecki M., Colonna M., Diamond M. S., Bhattacharya D. 2012. Spontaneous mutation of the Dock2 gene in Irf5-/- mice complicates interpretation of type I interferon production and antibody responses. Proc. Natl. Acad. Sci. U.S.A. 109: E898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fukui Y., Hashimoto O., Sanui T., et al. 2001. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature 412: 826 [DOI] [PubMed] [Google Scholar]
- 33. Nishikimi A., Fukuhara H., Su W., et al. 2009. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science 324: 384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanaka Y., Hamano S., Gotoh K., et al. 2007. T helper type 2 differentiation and intracellular trafficking of the interleukin 4 receptor-alpha subunit controlled by the Rac activator Dock2. Nat. Immunol. 8: 1067 [DOI] [PubMed] [Google Scholar]
- 35. Gotoh K., Tanaka Y., Nishikimi A., et al. 2010. Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J. Exp. Med. 207: 721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi G. P., Villadangos J. A., Dranoff G., et al. 1999. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 10: 197 [DOI] [PubMed] [Google Scholar]
- 37. Hardy R. R., Hayakawa K. 2001. B cell development pathways. Annu. Rev. Immunol. 19: 595 [DOI] [PubMed] [Google Scholar]
- 38. Naik S. H., Proietto A. I., Wilson N. S., et al. 2005. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 174: 6592 [DOI] [PubMed] [Google Scholar]
- 39. Savitsky D. A., Yanai H., Tamura T., Taniguchi T., Honda K. 2010. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc. Natl. Acad. Sci. U.S.A. 107: 10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paun A., Bankoti R., Joshi T., Pitha P. M., Stäger S. 2011. Critical role of IRF-5 in the development of T helper 1 responses to Leishmania donovani infection. PLoS Pathog. 7: e1001246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Negishi H., Ohba Y., Yanai H., et al. 2005. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. U.S.A. 102: 15989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richez C., Yasuda K., Watkins A. A., et al. 2009. TLR4 ligands induce IFN-alpha production by mouse conventional dendritic cells and human monocytes after IFN-beta priming. J. Immunol. 182: 820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lien C., Fang C. M., Huso D., Livak F., Lu R., Pitha P. M. 2010. Critical role of IRF-5 in regulation of B-cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 107: 4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang C. M., Roy S., Nielsen E., et al. 2012. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 13: 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Negishi H., Yanai H., Nakajima A., et al. 2012. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat. Immunol. 13: 659 [DOI] [PubMed] [Google Scholar]
- 46. Krausgruber T., Blazek K., Smallie T., et al. 2011. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12: 231 [DOI] [PubMed] [Google Scholar]
- 47. González-Navajas J. M., Lee J., David M., Raz E. 2012. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12: 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kayagaki N., Warming S., Lamkanfi M., et al. 2011. Non-canonical inflammasome activation targets caspase-11. Nature 479: 117 [DOI] [PubMed] [Google Scholar]
- 49. Kenneth N. S., Younger J. M., Hughes E. D., et al. 2012. An inactivating caspase 11 passenger mutation originating from the 129 murine strain in mice targeted for c-IAP1. Biochem. J. 443: 355 [DOI] [PMC free article] [PubMed] [Google Scholar]