Figure 2.
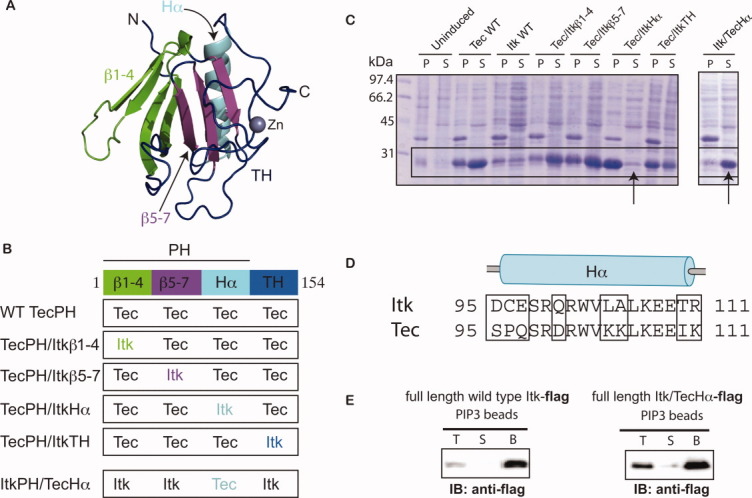
A chimeric approach based on the TecPH sequence reveals that the α-helix of ItkPH is responsible for its poor expression behavior. (A) Threaded structure of ItkPHTH onto the available BtkPHTH structures14, 15 (PDB: 1BTK and 1B55); threading was performed using I-TASSER;28 the ribbon tracing the protein backbone is colored to indicate the four regions used in construction of the Tec/Itk chimeras. β strands 1–4 are green, β strands 5–7 are purple, the α-helix is cyan, and the TH domain (or Btk motif) is dark blue. The structural model of ItkPH was visualized in PyMOL29 and is used in the subsequent figures. (B) Diagram describing the construction of the four Tec/Itk chimeras and the single Itk/Tec chimera. Coloring for each of the regions matches that described in (A). (C) Wild type (WT) and chimeric proteins were expressed in BL21(DE3) cells and analyzed for soluble expression; pellet (P) and soluble (S) fractions were analyzed by SDS-PAGE and the boxes indicate the bands corresponding to the PHTH constructs; the arrows point to the chimeras in which the α-helices of Tec and Itk have been swapped. (D) Sequence alignment of the alpha-helices of ItkPH and TecPH, showing the residues that were swapped in the helix chimeras (amino acids 95–111); boxes highlight the eight amino acid differences between Tec and Itk in this region. (E) Itk/TecHα retains canonical PIP3 binding activity in the context of full-length Itk; FLAG-tagged full-length wild-type Itk and full length Itk/TecHα are captured using PIP3-coated beads, washed, and analyzed by immunoblot (anti-FLAG): lanes correspond to total input (T), supernatant (S), and washed bead (B) samples.
