Figure 3.
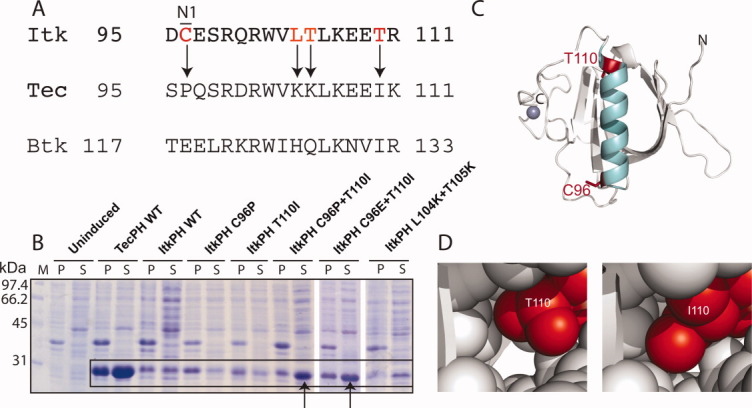
Minimal sequence changes in ItkPH required to produce soluble protein. (A) Sequence alignment of the α-helices of ItkPH, TecPH, and BtkPH, with arrows indicating the most significant amino acid differences that were targeted for mutation. It is interesting to note that position 96 is not conserved between mouse and human Itk sequences; mutation of the mouse sequence to that of human (C96R) did not alter the expression and solubility behavior of ItkPH (data not shown). (B) Soluble expression analysis of single and double helix mutations; arrows point to the C96P/T110I and C96E/T110I mutations that rescue the poor expression of wild type ItkPH. The overall expression level for wild-type ItkPH is quite low when compared with ItkPH C96P/T110I, ItkPH C96E/T110I, and TecPH. This suggests that expression of the wild-type ItkPH sequence leads to protein degradation rather than incorporation into inclusion bodies. (C) Structural model of the Itk PH-TH domain: α-helix shown in cyan, positions C96 and T110 shown in red, N- and C-termini are labeled and the Zn2+ ion coordinated by the TH domain is shown in blue. (D) Comparison of side-chain packing for threonine versus isoleucine at position 110 in the threaded model of ItkPH reveals a significant cavity for T110, whereas I110 packs tightly against adjacent side-chains.
