Figure 5.
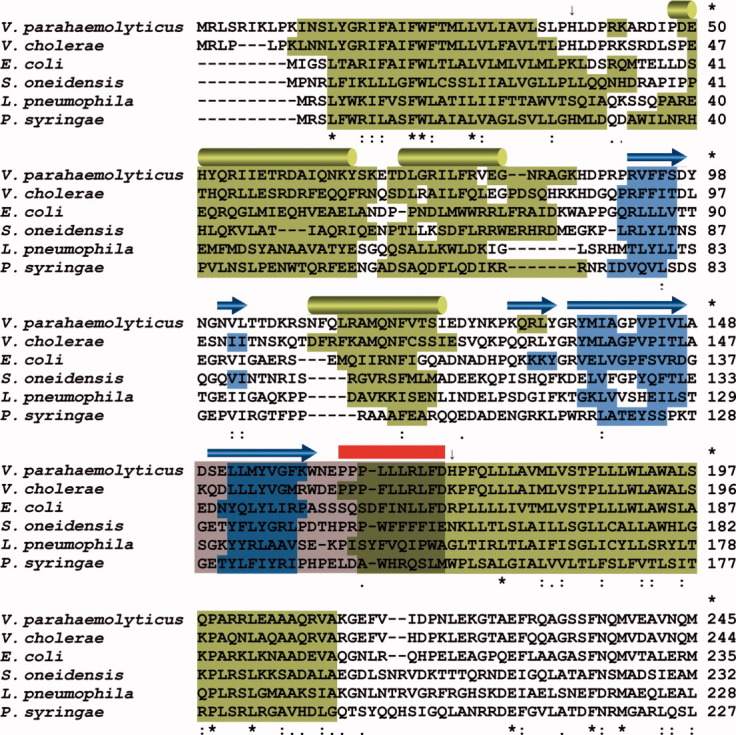
Multiple sequence alignment of CpxA proteins of γ-proteobacteria. Multiple sequence alignment of the CpxA proteins of V. parahaemolyticus, V. cholerae, E. coli, Shevanella oneidensis, Legionella pneumophila, and Pseudomonas syringae. Predicted α-helices and β-strands are highlighted on the sequence of each species in green and blue, respectively. α-helices (H1–H3) and β-strands (S1–S5) in Chain A of VpCpxA-peri are displayed in the first row as green cylinders and blue arrows, respectively. The C-terminal tail of VpCpxA-peri deleted for the NMR experiment is indicated by a red line, and the region predicted to play a role in E. coli CpxP binding is represented by a grey box.25 The N- and the C-terminal residues of VpCpxA-peri are indicated by arrows.
