Figure 1.
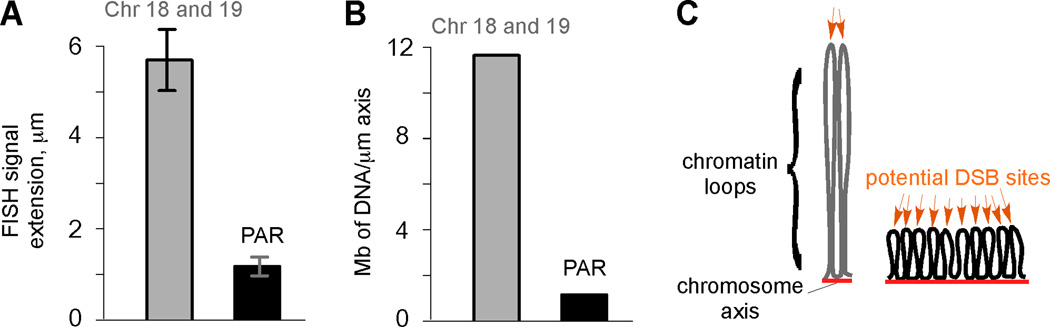
Higher-order structure of the PAR compared to autosomes. A) Measurements of the maximal distance of FISH signals from chromosome axes (average for subtelomeric chromosome 18 and 19 probes, gray bar; PAR probe, black bar) in cells at pachynema (mean ± SD). B) DNA content (Mb) per µm of chromosome axis in the subtelomeric regions of chromosomes 18 and 19, and the PAR. C) Model of how organization of DNA into chromatin loops can influence DSB frequency (after ref. 18). Only one homolog (each consisting of a pair of sister chromatids) is shown, with chromatin loops tethered to chromosome axes (red lines). Each chromatin loop is envisioned to provide an opportunity for meiotic DSB formation (orange arrows). DNA organized into more and smaller loops per Mb (right) presents the DSB machinery with more opportunities for break formation than if the same length of DNA is organized into fewer and larger loops (left).
