Abstract
This review summarizes the evidence indicating that mutagenic mechanisms in vivo are essentially the same in all living cells. Unique metabolic reactions to a particular environmental stress apparently target specific genes for increased rates of transcription and mutation, resulting in higher mutation rates for those genes most likely to solve the problem. Kinetic models which have demonstrated predictive value are described and are shown to simulate mutagenesis in vivo in E. coli, the p53 tumor suppressor gene, and somatic hypermutation. In all three models, direct correlations are seen between mutation frequencies and transcription rates. G and C nucleosides in single-stranded DNA (ssDNA) are intrinsically mutable, and G and C silent mutations in p53 and in VH framework regions provide compelling evidence for intrinsic mechanisms of mutability, since mutation outcomes are neutral and are not selected. During transcription, the availability of unpaired bases in the ssDNA of secondary structures is rate-limiting for, and determines the frequency of mutations in vivo. In vitro analyses also verify the conclusion that intrinsically mutable bases are in fact located in ssDNA loops of predicted secondary structures.
Keywords: mutagenesis, E. coli, p53, somatic hypermutation, kinetic models
1. Introduction
Due to the Unity of Biochemistry, first recognized by Kluyver in 1926 [1], evolution has bestowed a fundamental unity of biochemical behavior on all forms of life: the same substrates, enzymes, regulatory mechanisms, biosynthetic and catabolic pathways. The mechanism of mutagenesis, then, should be similar in microbes and man. However, this similarity is difficult to demonstrate because, in vivo, a system of coupled interacting biological components has properties far beyond those apparent from the analysis of these components in isolation. Thus, hierarchical schemes of control based on any single cellular event, such as “gene activation” or “enzyme-induction” is inadequate to account for biochemical behavior that necessarily owes its very existence to complex relationships among many kinds of events. Moreover, in vitro data may not be relevant to circumstances in vivo, because substrates are typically used at saturating levels while enzymes are used at low, rate-limiting concentrations in order to measure the rate of a reaction. In contrast, enzyme concentrations in vivo under steady-state conditions usually far exceed substrate concentrations, and therefore enzymes may not receive sufficient substrate to operate at maximum velocity. Thus, the amount of available substrate is rate-limiting [2–4]. Moreover, the effect of transcription on mutagenesis in vivo differs from circumstances typically used in vitro. For example, genotoxin-induced transcription of the p53 tumor suppressor gene in vitro has frequently been reported [5–7], but it is also known that transcription exposes ssDNA with G and C nucleotides that are intrinsically mutable [8,9]. Therefore, in vivo, the role of carcinogens in mutagenesis is confusing and difficult to predict, since mutagens both activate transcription and cause base damage (primarily G-to-T transversions) [10]. In spite of the fundamental importance and consequences of these insights concerning metabolism in vivo, these facts have been largely ignored, perhaps because further insights would likely require collaborations between biochemists and experts able to formulate kinetic models which simulate circumstances in vivo.
Our challenge is to understand these relationships in vivo. Fortunately, kinetic models have the unparalleled ability to cope with such complexity by calculating the consequences of an integration of the relevant information. As our biochemical knowledge grows, models will become increasingly essential for the integration and interpretation of this knowledge. They allow us to clarify which facts are compatible and pinpoint those that are not. Since kinetic models can simulate metabolism in living systems, they also provide a framework in which to judge the relevance of in vitro data to metabolism in vivo. However, models are only valid and useful if they have demonstrated predictive value. Kinetic models created by our computer program, mfg, have frequently demonstrated predictive value in the analyses of mutagenesis in three very different systems: Escherichia coli [11–15], the p53 tumor suppressor gene [10, 16], and in somatic hypermutation (SHM) during the immune response [17–20]. In these analyses, mutation frequencies were determined experimentally during transcription and during mfg-simulated transcription in order to predict: (1) the location of mutable bases in loops of high-stability secondary structures, and (2) the effects of transcription, supercoiling, and secondary structure stabilities on base mutability.
Thus, our research over the past ten years on the mechanisms underlying mutagenesis in E. coli, p53, and SHM is summarized and compared in order to determine whether the Unity of Biochemistry is, indeed, applicable to the mechanisms underlying mutagenesis in all forms of life.
2. Interactions of substrates, enzymes and stressors in vivo
One of the earliest investigators to use a simple kinetic model for finding critical variables in vivo was H. Kascer [21], who explained that “…it is necessary to uncover, both experimentally and logically the causal connections of a system without isolating the steps of which it is composed. The language in which such a system is described must of necessity be the language of molecular interactions, namely, kinetics. Our conventional logical apparatus, which is essentially a linear one and lacks quantitative rigor, cannot handle most of the situations which are of the essence of interacting systems. Some of the conclusions of the treatment which follows may therefore appear intuitively strange… but so much the worse for intuition”. His elegant insight into our need for kinetic models to understand metabolism in vivo prompted a brief description of his model, to help our readers grasp more complex models of mutagenesis.
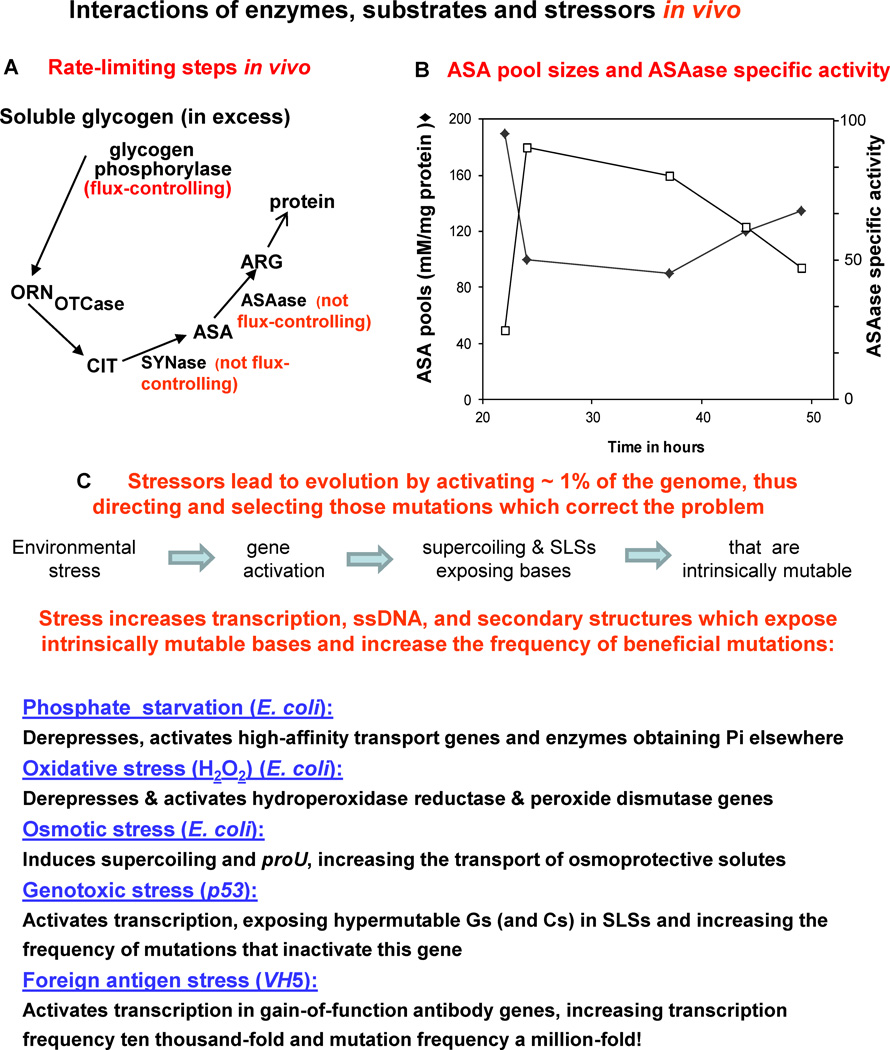
Kascer used the pathway of arginine synthesis in Neurospora to show, under steady-state conditions, the sensitivity of metabolite flux to changes in the level of an enzyme, depending upon its “kinetic position” in a metabolic sequence (Fig. 1A). Since glycogen is in excess, glycogen phosphorylase is rate-controlling for flux through the cycle, while CIT SYNase and ASAase are not. He used a simple mathematical model and five Neurospora mutants that varied in ASAase-specific activity, and he also measured the concentrations of the ASA pools and the ARG pools (not shown) (Fig. 1B). Thus, under steady-state conditions, Kascer correctly predicted: (1) no effect on ARG pool sizes in the five mutants because SYNase and ASAase are not rate-controlling; and (2) that changes in ASAase-specific activity would result in inverse correlations with ASA concentrations, which was also observed. Therefore, his model demonstrated predictive value: one enzyme, glycogen phosphorylase, is flux-controlling while two others, SYNase and ASAase, are not.
Fig. 1.
(A) Analyses of enzymes that are /are not rate-controlling under steady-state conditions in vivo. Soluble glycogen is in excess and therefore glycogen phosphorylase is flux-controlling, while two other essential enzymes, ASAase and SYNase, are not rate-controlling in vivo. (B) Kascer [21] used a simple mathematical model and five Neurospora mutants that varied in ASAase-specific activity, and also measured the concentrations of ARG and ASA. He correctly predicted no differences in ARG pool sizes (data not shown) as well as changes in ASAase-specific activity that resulted in inverse correlations with ASA concentrations. (C) Examples from both prokaryotes and eukaryotes in which stressors in the environment activate related genes, thus increasing the frequency of transcription and mutations that overcome the stress.
As illustrated in Fig. 1C, evolution usually occurs in response to stress, by activating relevant portions of the genome and selecting beneficial mutations that correct the problem [11, 12,22–24]. In the prokaryote E. coli, for example, starvation for inorganic phosphate de-represses the phoregulon, thus activating a new high-affinity phosphate transport system able to cope with lower phosphate levels, and also activates a hydrolase able to obtain phosphate from new sources [25]. Oxidative stress activates hydroperoxide reductase in E. coli [26], and the effect of osmotic stress on supercoiling and expression of the proU locus increases the transport of osmoprotective solutes [27]. In humans, genotoxic stress activates p53 transcription, thus exposing intrinsically hypermutable Gs and Cs in stem-loop structures (SLSs) [10,16,28], and foreign antigen stress also activates transcription, resulting in a striking increase in B cell mutation frequency [29–31]. Thus E. coli, the p53 tumor suppressor gene, and SHM in VH5 have apparently evolved similar solutions in response to environmental stressors. In each case, transcription provides the ssDNA that exposes unpaired intrinsically mutable bases, which are the essential precursors of mutation.
3. How to use our computer algorithm, mfg
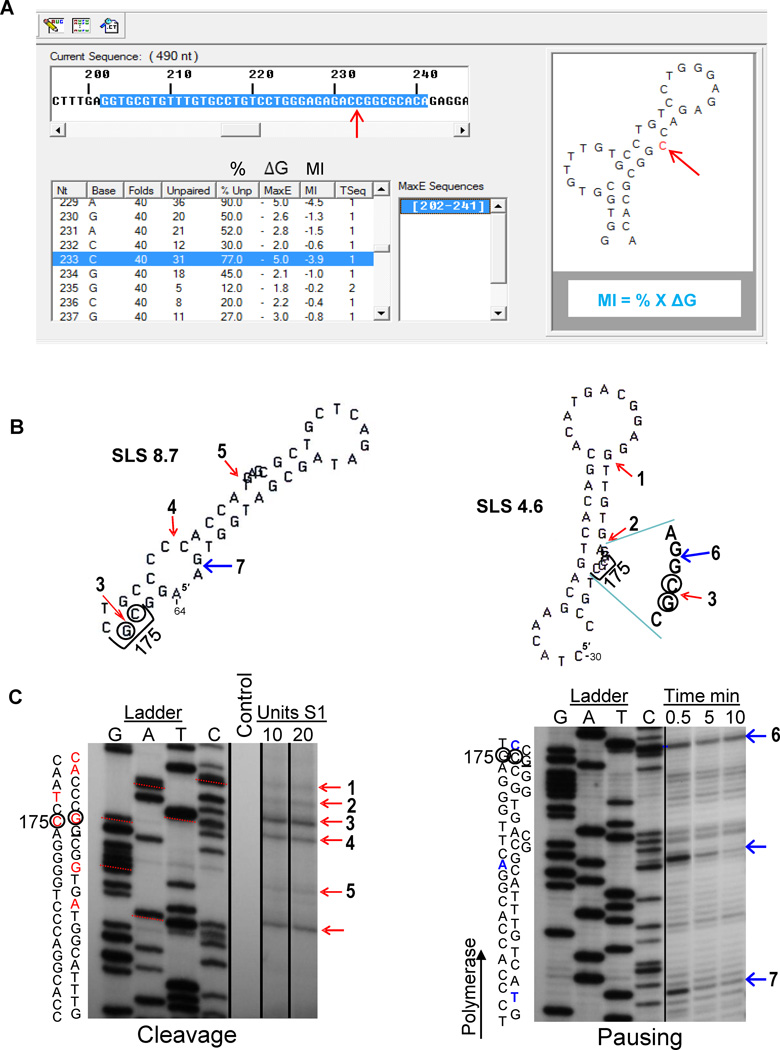
To simulate in vivo conditions in response to increased rates of transcription a computer algorithm, mfg, was developed [12]. During transcription the mutability of a base in a DNA secondary structure depends both on the stability of the structure and on the extent to which the base is unpaired. Mfg interfaces with the mfold program [32] which can fold single-stranded segments of a specified length for any given sequence and report all possible secondary structures that can form from each folded segment, in descending order of stability. As indicated in Fig. 2A, mfg uses mfold to analyze a sequence of ssDNA. The sequence and window size are first specified, and then mfold is instructed to fold the relevant successive sequences into SLSs (stem-loop structures). Mfg then reports the stability (Max E, or ΔG) of the most stable secondary structure in which a mutable base is unpaired, and also reports the percent of total folds in which it is unpaired. The Mutability Index (MI) of each unpaired base is the product of these two variables, i.e. MI = % unpaired X ΔG, or MI = % X ΔG (Fig. 2A). Two such structures are shown in Figure 2B, namely, SLS 8.7 and SLS 4.6. The website http://www.dbs.umt.edu/research_labs/wrightlab/upload/mfg.html provides directions for using mfg to analyze DNA secondary structures formed during transcription. An example of mfg output is shown in Figure 2A. A segment of ~ 400 nts in the non-transcribed strand of the selected sequence is first pasted into the mfg program. A frame length for viewing the subsequent successive secondary structures is then chosen, and these segments are copied to the clipboard and loaded in to the mfold program http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold for folding. Once the folded sequences are available they are loaded back into mfg by selecting “save target as” from the dropdown menu. The successive sequences are then saved to an mfg-associated file. This ct file is processed by mfg, which selects subsequences where subsequent bases are unpaired, and then gives the frequency (%) with which they are unpaired and the stability (ΔG, or Max E) of the most stable structure in which they are unpaired. As seen by the computer output (Fig. 2A), these data are shown along with the bases in the sequence. Thus, by selecting any base, the highest energy structure in which that base is unpaired is displayed, with the selected base highlighted in blue.
Fig. 2.
(A) Read out from the mfg computer program highlighting the sequence that is folded in the top window. Folding data are shown on the lower left (Note: MaxE is synonymous with ΔG) and the structure generated is shown to the right, exposing the mutable C (arrowed in red). (B) Selected p53 structures associated with the sequence surrounding “hot spot” codon 175 together with (C) accompanying gels showing cleavage sites from S1 endonuclease digestions of supercoiled plasmid DNA and pausing sites from T7 DNA polymerase. Structures reflect folding of the non-transcribed strand, while experimental cleavage and pausing data were derived from the transcribed strand. Two, 44-nucleotide structures, SLS 8.7 and SLS 4.6, are shown from an mfg analysis of exon 5 (red arrows indicate experimentally-determined cleavage sites and blue arrows indicate polymerase pausing sites; numbers 1–7 correspond to sites in gels below). For both gels, the sequence (transcribed strand) is indicated on the left, hypermutable bases are circled, and the third position of codon 175 is underlined. The unnumbered cleavage and pause site arrows correspond to high-stability structures published elsewhere [10].
Cleavage: An autoradiograph of a denaturing polyacrylamide gel that contains the sequence in the exon 5 region (ladder), shown juxtaposed to S1 nuclease-digested dsDNA plasmid fragments using the same primer. Lane 1 is the negative control; lanes 2 and 3 show reactions using 10U and 20U S1, respectively. Corresponding bases involved in cleavage are indicated in the sequence in red. Red arrows indicate experimentally determined S1 cleavage sites, where numbers correspond to loops in structures shown. (Adapted from [10]).
Pausing: An autoradiograph of a denaturing polyacrylamide gel containing the exon 5 ladder, and three time points corresponding to T7 DNA polymerase exposure. Polymerase pauses are indicated by bands (arrowed blue) and corresponding bases in the sequence are shown in blue. Arrow numbers correspond to bases of stems in the structures shown above. (Done as previously described [19], using the primer- 5′-CTAAGAGCAATCAGTG-3′).
To our knowledge the mfg program is unique for predicting the successive formation and inter-conversion of the most stable SLSs that presumably exist in vivo during transcription of sequences hundreds of nucleotides long. This algorithm is based upon logical assumptions: Each successive fold must be initiated by an unpaired base, and the most stable structure in which each base is unpaired will dominate the folding pathway. Thus, the mfg program has allowed us to discover and describe a here-to-for unknown mechanism of mutagenesis that incorporates and augments the underlying causes of transcription-driven SLSs containing intrinsically mutable unpaired bases [11, 24]. This program has recently been used in the analysis of viral RNA secondary structures [33], and in investigations to determine the extent to which mutations occur in unpaired secondary structures of the E. coli glycerol kinase glpK 218 allele [15]. Kim et al. [14] have used mfg for analyses of transcription-associated mutations, and suggest that the genome sequence has evolved to increase protein evolvability under stress. In mfg analyses of human rearranged IGHV3-2301 gene sequences Duvvuri et al. [20] found that hypermutable bases were located in the ssDNA of stable SLSs.
4. Mfg-predicted secondary structures exist
In order to confirm mfg-analyses indicating that highly mutable Gs and Cs in p53 are actually located in ssDNA loops of predicted SLSs, plasmid DNA containing codon 175 in exon 5 of p53 was analyzed using S1 endonuclease (Fig. 2B and C). Selected p53 structures associated with the sequence surrounding “hot spot” codon 175 and accompanying gels shows cleavage sites from S1 endonuclease digestions of supercoiled plasmid DNA and pausing sites using T7 DNA polymerase. The SLSs are from the non-transcribed strand, while experimental cleavage and pausing data were derived from the transcribed strand. Comparable analysis demonstrating the existence of predicted SLSs in p53 have been demonstrated for hypermutable codons 245, 248, 273, and 282 [10], and in somatic hypermutation [19] pausing analysis demonstrated the existence of three different sized segments of VH5.
5. G and C bases are intrinsically mutable
Table 1 summarizes the characteristics and mutability of nucleosides (predominately Gs and Cs) in ssDNA, demonstrating their intrinsic instability and fate. In particular, unpaired Gs primarily mutate to A and Cs mutate to T. Compelling evidence for the mechanism underlying base instability is shown by the three examples of intrinsic mutability, i.e., of nucleosides and two silent mutations, which can only reflect intrinsic base instabilities since they do not alter the encoded amino acid and are not selected [see 16,34,35]. These data underlie key questions that we address below: Is the availability of intrinsically mutable unpaired bases via transcription rate-limiting for mutagenesis? If so, does this fully explain the mechanistic link between the frequencies of transcription and mutation in E. coli auxotrophs, p53 exons, and SHM? As demonstrated by Kascer, kinetic models are essential tools for simulating such complex relationships in vivo in order to answer these fundamental questions.
Table 1.
Correlations between stability and mutability of G and C bases in loops of SLSs
| G-to-A | G-to-T | G-to-C | C-to-T | C-to-A | C-to-G | |
|---|---|---|---|---|---|---|
| Intrinsic: Thermodynamic characteristics of nucleosides leading to mutation in ssDNA | ||||||
| % of total Mutations | ~95% a | na | na | 95.6% | ~2.2% | ~2.2% |
| Selected: Background mutations in E. colilacI gene b | ||||||
| % of total Mutations | 53.7% | 12.9% | 33.3% | 62.1% | 22.1% | 15.8% |
| Selected: Missense mutations in eight codons of p53c | ||||||
| % of total Mutations | 75.9% | 20.1% | 4.0% | 93.0%d | 1.5% | 5.5% |
| Intrinsic: Silent mutations in all exons of p53d | ||||||
| % of total Mutations | 78.4% | 11.5% | 10.1% | 79.3% | 13.7% | 7.0% |
| Intrinsic: Silent mutations in VH framework regions e | ||||||
| % of total Mutations | 67.5% | 16.0% | 16.5% | 80.4% | 7.8% | 11.8% |
| Selected: Missense germline light chain mutations e | ||||||
| % of total Mutations | 72.7% | 7.6% | 19.7% | 81.8% | 15.2% | 3.0% |
About 5,000 purine bases turn over each day due to hydrolytic depurination, with G usually being replaced by A because of its size [37].
lacI mutations from Isbell and Fowler [36].
In p53, hypermutable Cs are methylated and mutate directly to T [16].
Silent mutations are presumed to be under-reported in the IARC TP53 database [38] because they do not result in cancer.
Data were obtained from VH5, VH94, and VH186.2 genes [39].
6. Correlations between mutation frequencies, base mutability and loops in SLSs
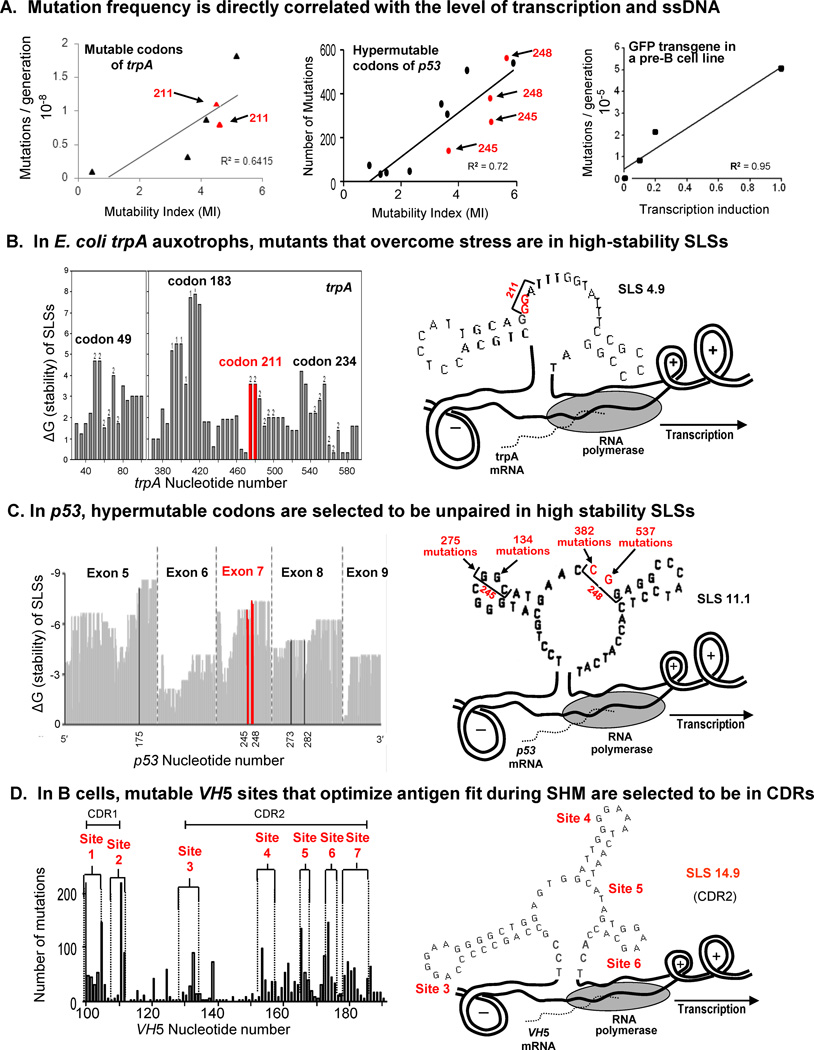
As shown in Fig. 3A, direct correlations between mutation and transcription frequencies occur in both prokaryotes, such as a trpA E. coli auxotroph [12,36] and eukaryotes, e.g., the p53 tumor suppressor gene, [10,16], and a pre-B cell line [29,30]. In each case, the mutable bases are located in loops of identified secondary structures, as demonstrated in Fig. 3B and C for E. coli and p53, respectively. In Fig. 3B, comparisons are made between the stability (ΔG) of four trpA codons and the location of two mutable Gs in codon 211 of SLS 4.9. Fig. 3C shows that Exon 7 of p53 has two of the most hypermutable codons, 245 and 248, which are located as shown in SLS 11.1. In the heavy chain variable region VH5 gene (Fig. 3D) mutable bases have been identified in predicted mutable sites 1–7 in complementarity-determining regions 1 and 2 (CDR1 and CDR2). The location of four of these sites (3 to 6) is shown in loops (unpaired regions) in the predominant secondary structure, SLS 14.9.
Fig. 3.
(A) Correlations between mutation frequencies and rates of transcription in mutable codons of trpA, in hypermutable codons of p53, and in a GFP transgene in a pre-B cell line. (B) Mutants of E. coli trpA auxotrophs located in a loop of SLS 4.9 [12,36]. Mutable codon 211 is highlighted in red to indicate its stability and location in SLS 4.9. (C) Hypermutable codons in exon 7 of p53 are shown to be exposed in loops of the high-stability SLS 11.1 [10, 16]. (D) During SHM codons are primarily located in the CDRs, and hypermutable codons 3–6 are located in loops of the dominant secondary structure, SLS 14.9 [29,30].
7. Somatic hypermutation
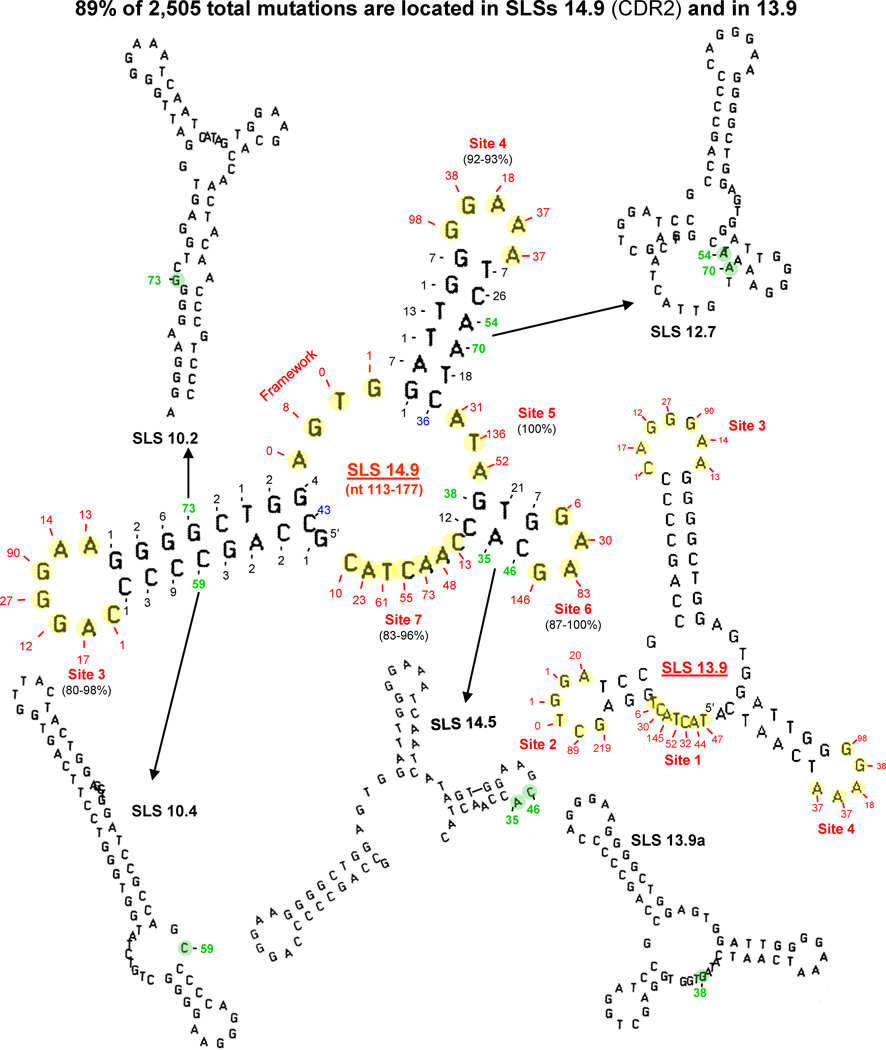
The mechanisms underlying the immune response in pre-B cell lines are fascinating. A 10,000-fold increase in transcription initiates, and is mechanistically linked to a million-fold increase in mutation frequency [31], especially during phase 1 of SHM, predicted to follow B cell activation [40]. Moreover, there is a direct correlation between increasing levels of transcription and mutation frequency [29,30]. Extremely high frequencies of mutation are apparently essential for producing the number of variations required to test, change and coordinate mutations in fitting antibody to antigen. The rate of VH and VL transcription is negligible in germ-line DNA, but variable-region gene rearrangements close the gap between enhancers and promoters to increase the rate of transcription by ~10,000-fold. Polymerase pausing analyses (not shown, but similar to those shown in Fig. 2B and C) of three different-sized segments of VH5 in the non-transcribed strand have verified the existence of the two major mfg-predicted 65 nt secondary structures seen in Fig. 4 (SLS 14.9) and (SLS 13.9) [19]. The present model of SHM accounts for all seven mutable sites in ssDNA loops in the variable region, and 89% of the 2,505 total mutations identified are accounted for in unpaired bases of SLSs 14.9 and 13.9.
Fig. 4.
The current model of VH5 in B cells (see Fig. 3D) in which 89% of 2505 mutations in CDR1 and CDR2 have been found to exist as unpaired bases (shown highlighted in yellow) in loops of the predominant 65-nt structures of SLS 14.9 and SLS.13.9. The numbers of mutations [39] are indicated at each yellow base. Mutations occurring in sites 1–7 are shown in red, while those in lower-stability structures are shown in green. Two mutable bases at the base of stems are shown in blue.
8. Rate-limiting variables for mutagenesis
The intrinsic instability of G and C nucleosides in ssDNA (Table 1), as well as the high number of G and C silent mutations in all exons of p53 (78.4% Gs and 79.3% Cs), and of VH framework regions (67.5% Gs and 80.4% Cs) provide compelling evidence for an intrinsic mechanism of mutability, since the outcome of these mutations does not alter the encoded amino acid and is not selected. The direct correlation (Fig. 3A) between mutation and transcription frequencies in an E. coli auxotroph, the hypermutable codons of p53, and in a pre-B cell line also implicates ssDNA as the essential rate-limiting substrate for mutagenesis.
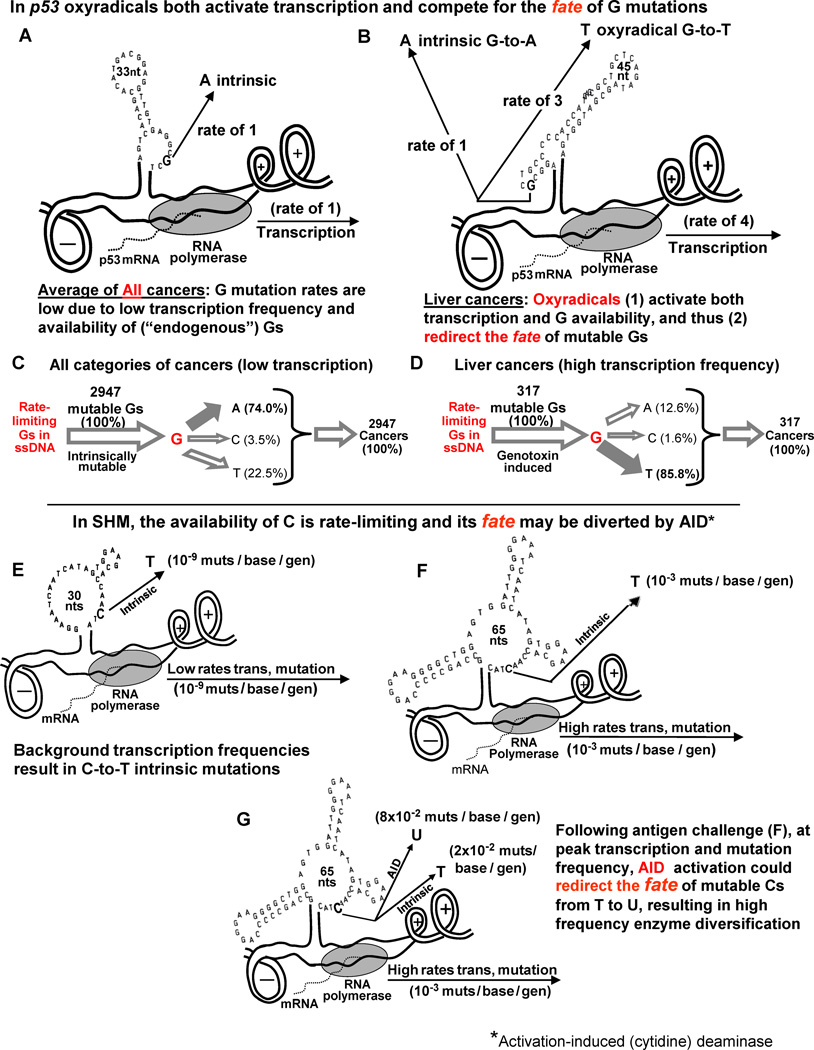
Kinetic models shown in Fig. 5A-D help to clarify a key question with respect to liver cancers induced by genotoxins and involving p53: In liver cancers, is the availability of unpaired intrinsically mutable Gs in ssDNA rate-limiting for mutation frequency? The answer is yes! Circumstances in vivo at low “endogenous” levels of transcription are depicted in Fig. 5A and C, showing that the majority of intrinsic G mutations (74.0 %) are to A, and that the availability of Gs in ssDNA is rate-limiting for mutation frequency (see Supp Table 2 of [10]). Fig. 5B and D describe the dual effects of oxyradicals, which both activate transcription (about 4 fold) and increase G-to-T transversions (to 85.8%), compensated for by decreases in G-to-A and C mutations (Fig. 5D). Thus, oxyradicals compete for the fate of rate-limiting G mutations. In other words, since Gs in ssDNA are rate-limiting for mutation frequency, an increase in G-to-T transversions must be compensated for by decreases in G-to-A and C mutations [10].
Fig. 5.
Kinetic models of the fate of intrinsically mutable Gs in p53 and Cs in SHM. Top Half: (A and C) The frequency of G mutations in All cancers (at background levels of transcription in the absence of mutagens) is low, due to the low frequency of transcription in which the majority of mutations (74.0 percent) are G-to-A (intrinsic) compared with liver cancers. (B and D) Oxyradicals activate transcription (about 4-fold) as well as G availability, thereby diverting most of the G mutations (85.8%) from A to T, as depicted in (D). Thus, the relative rate of G-to-A intrinsic mutations is low. Cancer frequency data were obtained from [38].
Bottom Half: Kinetic models of circumstances resulting in mutation frequencies in VH5 during SHM. (E) Low background frequencies of about 10-9 mutations/base/generation resulting in C-to-T intrinsic mutations. (F) The consequences of the ~10,000-fold increase in transcription frequency in response to antigen challenge targeted primarily at Cs to T in ssDNA. (G) Predicted events occurring at the peak of Phase 1 of SHM as the result of AID activation and a switch from high frequency C-to-T mutations to high frequency C-to-U mutations, leading to enzyme diversification.
As depicted in Fig. 5E-G, similar compensatory shifts in the fate of rate-limiting Cs occur during SHM. Fig. 5E models background levels of transcription and shows intrinsic C-to-T mutations occurring at a frequency of ~10−9 mutations/base/generation. Foreign antigen challenge (Fig. 5F) results in a striking increase in transcription frequency, from 10−9 to 10−3 mutations/base/generation, and a roughly million-fold increase in mutation frequency [31]. The role of activation-induced (cytidine) deaminase (AID) has not yet been entirely clarified, and we suggest that its activation may in part serve to redirect the fate of mutable Cs from T to U (Fig. 5G), leading to high frequency enzyme diversification compensated for by lower rates of intrinsic C-to-T mutations [19].
9. Conclusions and perspective
As demonstrated by the work of Kascer [21], in vivo models can provide conceptual insights that are not intuitively obvious and help to reveal which variables are, and which are not rate-controlling, depending upon their “kinetic” position in a metabolic sequence. In understanding the mechanisms underlying mutagenesis, perhaps the most difficult conceptual insight that is not intuitively obvious is the intrinsic instability and mutability of G and C bases (Table 1), and the fact that they are rate-limiting for mutagenesis during transcription (Fig. 5C and D).
Models created by using our computer program, mfg, have demonstrated predicted value in the analyses of mutagenesis in microorganisms [12–15,24], and in p53 [10,16], by: (a) correlations between predicted and experimentally-determined mutation frequencies during transcription, (b) the predicted effects of transcription, promoter strength and supercoiling on base mutability, and (c) in vitro analyses that confirm the location of the twelve most mutable bases in predicted ssDNA loops of secondary structures. The predictive value of our mfg SHM model [17–19] has also been demonstrated in several ways by: (a) the location of high mutation frequency bases in mfg-predicted highly exposed unpaired bases, (b) the fact that unpaired bases in SLS 14.9 are identical to those identified independently in ssDNA patches during transcription [41], (c) the fact that the great majority of mutations in the non-transcribed strand occur within loops of predicted high-stability SLSs, (d) the observation that 89% of the 2,505 mutations reported in SLS sites 1–7 of these regions are predicted by mfg to occur in identified loops of secondary structures in the encoded CDR, and (e) in vitro analyses that have verified the existence of SLS 14.9.
Given the proposed model of mutagenesis in E. coli, p53 and SHM, the rate of transcription plays a dominant role in mutation frequency, and any method of down-regulating the rate of p53 transcription, for example, to baseline levels under stressful conditions may lower the incidence of lung or liver cancers.
Acknowledgements
We thank William Knight for creating mfg, Aaron Hunt for his insight into the use of mfg, and Dennis Reschke for his expertise in pausing and cleavage experiments.
Funding This work was supported by the Stella Duncan Memorial Research Institute and by the NIH grant R01 CA099242.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kluyver AJ, Donker HJL. Die Einheit in der Biochemie. Chem d Zelle u Gew. 1926;13:134–190. [Google Scholar]
- 2.Krebs H. Control of metabolic processes. Endeavour. 1957;16:125–132. [Google Scholar]
- 3.Srere PA. Enzyme concentrations in tissues. Science. 1967;158:936–937. doi: 10.1126/science.158.3803.936. [DOI] [PubMed] [Google Scholar]
- 4.Sauer F, Erfle JD, Binns MR. Turnover rates and intracellular pool size distribution of citrate cycle intermediates in normal, diabetic and fat-fed rats estimated by computer analysis from specific activity decay data of 14C-labeled citrate cycle acids. Eur J Biochem. 1970;17:350–363. doi: 10.1111/j.1432-1033.1970.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 5.Hellin AC, Calmant P, Gielen J, Bours V, Merville MP. Nuclear factor – kappaB-dependent regulation of p53 gene expression induced by daunomycin genotoxic drug. Oncogene. 1998;16:1187–1195. doi: 10.1038/sj.onc.1201638. [DOI] [PubMed] [Google Scholar]
- 6.Pei XH, Nakanishi Y, Takayama K, Bai F, Hara N. Benzo[a]pyrene activates the human p53 gene through induction of nuclear factor kappaB activity. J Biol Chem. 1999;274:35240–35246. doi: 10.1074/jbc.274.49.35240. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Shimizu H, Yamamoto K. Identification of a novel p53 promoter element involved in genotoxic stress-inducible p53 gene expression. Mol Cell Biol. 1995;15:4489–4496. doi: 10.1128/mcb.15.8.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 9.Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright BE, Schmidt KH, Hunt AT, Lodmell JS, Minnick MF, Reschke DK. The roles of transcription and genotoxins underlying p53 mutagenesis in vivo. Carcinogenesis. 2011;32:1559–1567. doi: 10.1093/carcin/bgr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright BE. Mini Review, A biochemical mechanism for nonrandom mutations and evolution. J Bacteriol. 2000;182:2993–3001. doi: 10.1128/jb.182.11.2993-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt KH, Reimers JM, Wright BE. The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol Microbiol. 2006;60:1251–1261. doi: 10.1111/j.1365-2958.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Lee BS, Tomita M, Kanai A. Transcription-associated mutagenesis increases protein sequence diversity more effectively than does random mutagenesis in Escherichia coli. PLoS One. 2010;5:5–12. doi: 10.1371/journal.pone.0010567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Applebee MK, Joyce AR, Conrad TM, Pettigrew DW, Palsson BØ. Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli. J Biol Chem. 2011;286:23150–23159. doi: 10.1074/jbc.M110.195305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright BE, Reimers JM, Schmidt KH, Reschke DK. Hypermutable bases in the p53 cancer gene are at vulnerable positions in DNA secondary structures. Cancer Res. 2002;62:5641–5644. [PubMed] [Google Scholar]
- 17.Wright BE, Schmidt KH, Minnick MF, Davis N. I. VH gene transcription creates stabilized secondary structures for coordinated mutagenesis during somatic hypermutation. Mol Immunol. 2008;45:3589–3599. doi: 10.1016/j.molimm.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright BE, Schmidt KH, Davis N, Hunt AT, Minnick MF., II Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol Immunol. 2008;45:3600–3608. doi: 10.1016/j.molimm.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright BE, Schmidt KH, Hunt AT, Reschke DK, Minnick MF. Evolution of coordinated mutagenesis and somatic hypermutation in VH 5. Mol Immunol. 2011;49:537–548. doi: 10.1016/j.molimm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duvvuri B, Wu GE. Gene Conversion-Like Events in the Diversification of Human Rearranged IGHV3-23*01 Gene Sequences. Front Immunol. 2012;3:158. doi: 10.3389/fimmu.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kascer H. In: Biological Organization at the Cellular and Super-cellular level. Harris RJC, editor. New York, NY: Academic Press Inc; 1963. pp. 25–41. [Google Scholar]
- 22.Pybus C, Pedraza-Reyes M, Ross CA, Martin H, Ona K, Yasbin RE, Robleto E. Transcription-associated mutation in Bacillus subtilis cells under stress. J Bacteriol. 2010;192:3321–3328. doi: 10.1128/JB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright BE. Micro Review, Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- 25.Makino K, Amemura M, Kim SK, Nakata A, Shinagara H. Mechanism of transcriptional activation of the phosphate regulon in Escherichia coli. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C.: ASM Press; 1994. pp. 5–12. [Google Scholar]
- 26.Storz G, Jacobson FS, Tartaglia LA, Morgan RW, Silveira LA, Ames BN. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli : genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S.typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 28.Murata J, Tada M, Iggo RD, Sawamura Y, Shinohe Y, Abe H. Nitric oxide as a carcinogen: analysis by yeast functional assay of inactivating p53 mutations induced by nitric oxide. Mutat Res. 1997;379:211–218. doi: 10.1016/s0027-5107(97)00149-8. [DOI] [PubMed] [Google Scholar]
- 29.Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 30.Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 31.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, Low DE, Gubbay JB. Genetic variability of human respiratory syncytial virus a strains circulating in ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer B, Kusmierek JT. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- 35.Schaaper RM, Dunn RL. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isbell RJ, Fowler RG. Temperature-dependent mutational specificity of an Escherichia coli mutator, dnaQ49, defective in 3'----5' exonuclease (proofreading) activity. Mutat Res. 1989;213:149–156. doi: 10.1016/0027-5107(89)90146-2. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 38.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 39.Zheng NY, Wilson K, Jared M, Wilson PC. Intricate targeting of immunoglobulin somatic hypermutation maximizes the efficiency of affinity maturation. J Exp Med. 2005;201:1467–1478. doi: 10.1084/jem.20042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 41.Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]