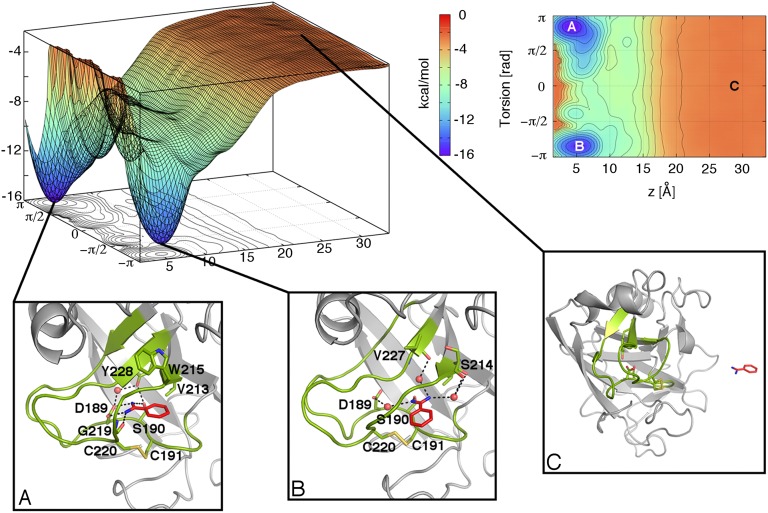
Fig. 2.
The FES of the benzamidine/trypsin binding is computed using a reweighting algorithm (50) as a function of the projection on the z axis of the ligand center of mass and a torsion CV, where z is the axis of the funnel (Table S1). Isosurfaces are shown every 1 kcal/mol. (Insets A–C) Conformations representing the bound poses, basin A (Inset A) and basin B (Inset B). C shows one of the isoenergetic conformations representing the unbound state. In the unbound state, z > 20 Å, the ligand can assume a large number of orientations, which are represented by states with similar energy values, as shown in the FES.