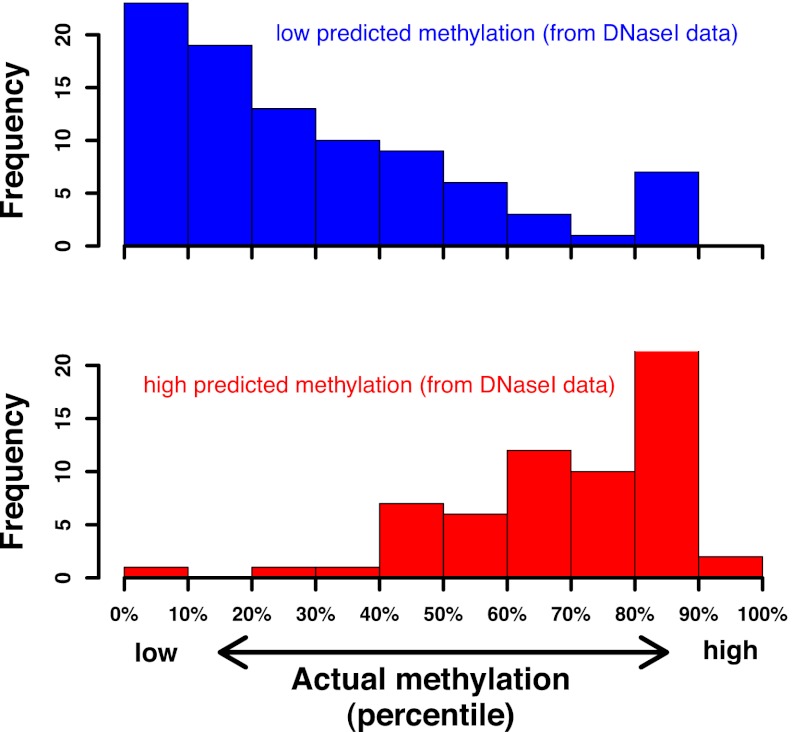
Fig. 4.
Genomic methylation status can be predicted from dense in vivo DNase I footprints. Starting from in vivo DNase I footprinting data for the IMR90 cell line, a set of nonoverlapping windows (2,500 bp long) containing at least 400 cleavage events with hexamer context NNNpCGN was identified. Next, for each window, the observed number of cleavages upstream of CpG dinucleotides was compared with the expected number. This allowed us to infer the methylation status of the corresponding DNA. To validate our predictions, we ranked all windows by their actual degree of methylation as measured by Lister et al. (36). Shown is the distribution of ranks for the subset of windows predicted to be hypomethylated (red) and hypermethylated (blue), respectively.