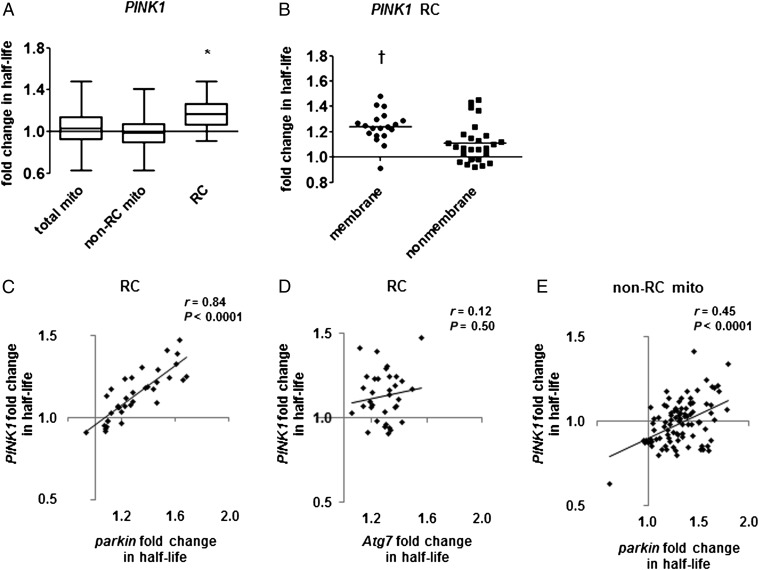
Fig. 3.
PINK1 null mutants have a selective impairment of RC protein turnover. (A) PINK1 null mutation prolongs the mean half-life of RC proteins but not other mitochondrial proteins. Box-and-whisker plots of fold change in half-life show median, quartiles, and extreme values. Mean fold change: total mito = 1.04 ± 0.16 (n = 147 proteins; P = 0.082 by nested ANOVA), non-RC mito = 0.99 ± 0.13 (n = 102), RC = 1.17 ± 0.16 (n = 45). *P < 0.0005, mutant vs. control, by nested ANOVA. (B) Mutation in PINK1 has a larger effect on membrane-bound than on nonmembrane RC subunits (mean fold change = 1.24 ± 0.13 vs. 1.11 ± 0.16). Horizontal lines indicate the median. †P < 0.005 by t test. (C and D) The effects of PINK1 mutation on RC protein half-lives strongly correlate with (C) the effects of mutation in parkin but not (D) the effects of mutation in Atg7 (n = 36 for parkin; n = 34 for Atg7). (E) The effects of PINK1 mutation on the half-lives of non-RC mitochondrial proteins correlate significantly with the effects of parkin mutation (n = 94). The regression line has a negative y intercept, suggesting a uniform shift to faster mitochondrial protein turnover in PINK1.