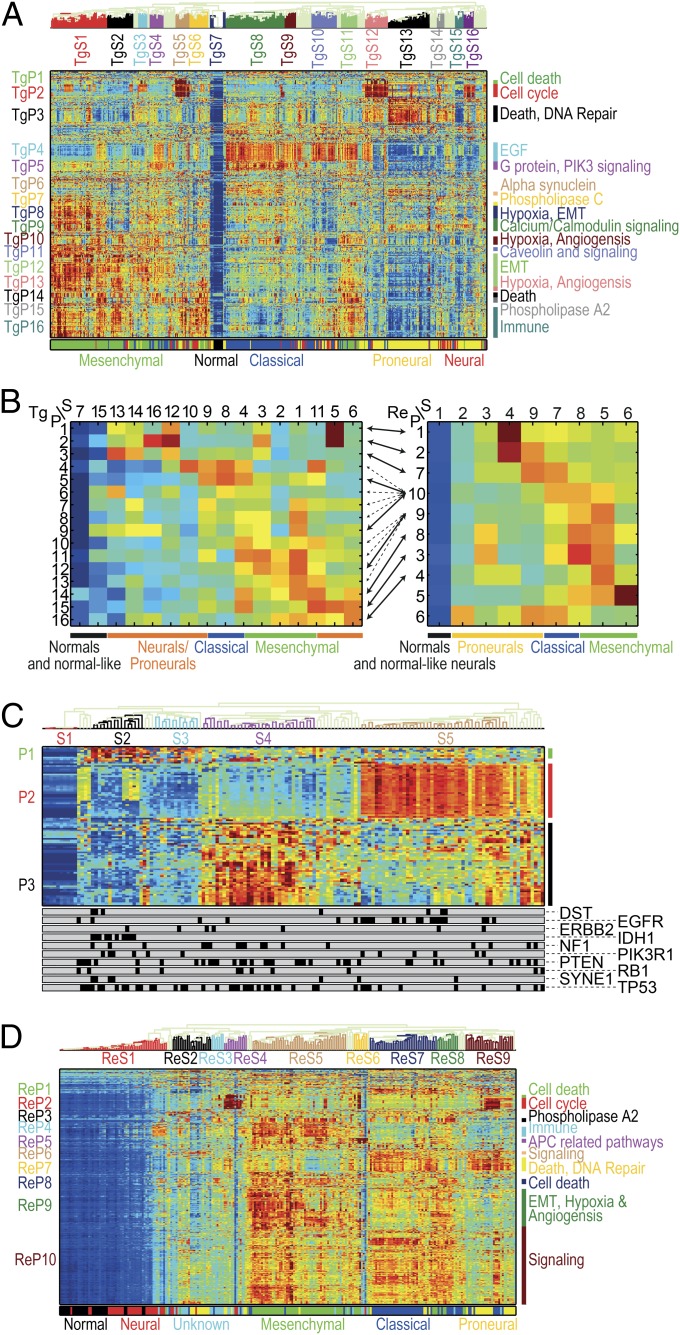
Fig. 2.
(A) Pathway deregulation scores (PDSs) of the TCGA glioblastoma (GBM) dataset (17). Each row corresponds to a pathway and each column to a sample. Pathways and samples are clustered according to PDS. Blue color represents low score (“no deregulation”) and red high. The bottom bar represents the GBM subtype. Notice that pathway-based clustering captures the subtypes well and identifies a secondary substratification. (B) Summary of clustered PDS for the TCGA (Left) and REMBRANDT (19) (Right) GBM datasets. Each row corresponds to a pathway cluster and each column to a sample cluster, displaying the median value of deregulation for each pair of clusters. Arrows connect between pathway clusters that match (that is, the pathways in the clusters have significant overlap). When several matches are significant (as for ReP9 and ReP10) all are shown in dashed arrows, except for the extremely significant ones (P < 10−5). Some of the neurals/proneurals are mostly not deregulated, and some are deregulated on TgP1–TgP3 or matching ReP1/ReP2/ReP7. Classical tumors are deregulated on TgP4/TgP5 and possibly TgP6/TgP7 as well as matching ReP10 (and unmatched ReP6/ReP7). Mesenchymal samples are highly deregulated on TgP8–TgP16 as well as matching ReP8–10/ReP3/ReP4 (and unmatchable ReP5). The classical-mesenchymal cluster TgS4 matches ReS8, and indeed they are both deregulated on TgP4/TgP5/TgP10–12/TgP14/TgP15 and matching ReP8–10/ReP3 (as well as unmatchable ReP5). (C) Normalized PDS of 94 pathways correlated with mutations. The bottom bars display the mutation status, each bar for one gene (samples with mutation are marked in black). Cluster S1 corresponds to normal samples, S2 mostly to samples with IDH1 mutations, S4 mostly to samples with NF1 mutations, and S5 mostly to samples with EGFR mutations. Notice pathway cluster P2, which consist mostly of EGF-activated pathways, and is highly deregulated on the EGFR mutated samples. (D) Normalized PDS of the REMBRANDT GBM dataset. As in A, the pathway clusters correspond to the known subtypes but offer additional substratifications.