Figure 4.
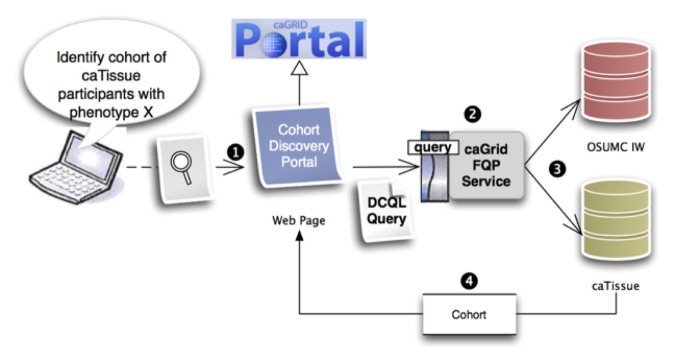
Overview of tissue cohort discovery tool implementation, in which: 1) end users pose a query via a cohort discovery portal built as a derivative of the caGRID portal platform; 2) that query is distributed and executed using Distributed Common Query Language (DCQL) via a TRIAD-specific instance of the caGrid-developed Federated Query Processor (FQP); 3) the ensuing source-specific queries, as specified via the initial DCQL statement and related semantic metadata and object modes, is executed against source systems; and 4) aggregate cohort-specific result sets are communicated to the portal interface and presented to the end user from FQP. In this example instance, phenotype data is being retrieved from the OSUMC IW, and biospecimen management data is being retrieved from a project-specific instance of caTissue Suite.
