Fig. 6.
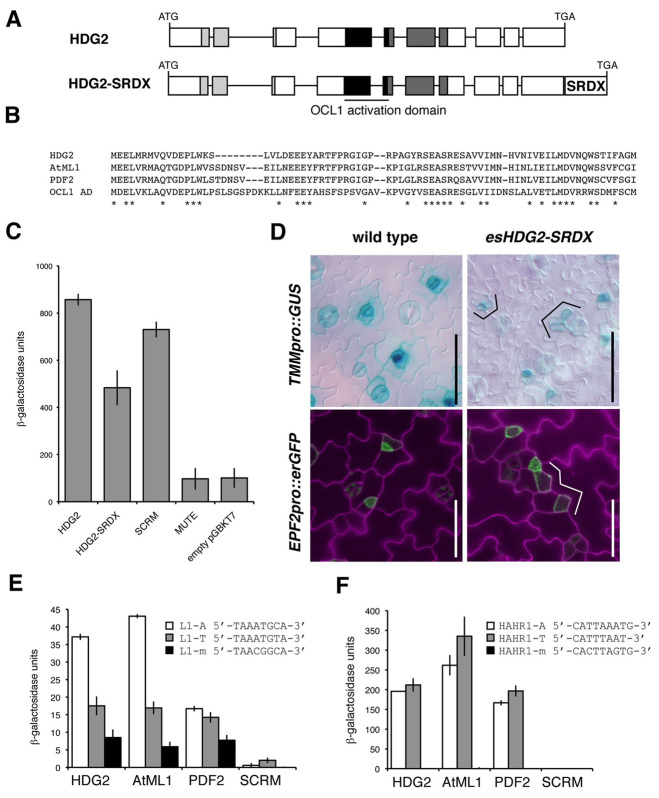
HDG2 is a transcriptional activator that can bind to both L1 box and HAHR1 box. (A) The HDG2 gene, from translational initiation codon to termination codon. Boxes indicate exons; lines indicate introns; light-gray boxes indicate homeodomain; dark-gray boxes indicate START domain, which also contains a transcriptional activation domain of maize OCL1 (black boxes). HDG2-SRDX has additional 12 amino acids (LDLDLELRLGFA) for forced gene repression. (B) Sequence similarity of HDG2 with AtML1, PDF2 and OCL1 activation domain region. (C) Transcriptional autoactivation of reporter lacZ gene in yeast by HDG2. Addition of SRDX reduces autoactivation. SCRM and MUTE, a known transcriptional activator and repressor, respectively (Kanaoka et al., 2008), were used as positive and negative controls. (D) Induced overexpression of HDG2-SRDX confers an epidermal phenotype with delayed stomatal precursors, resulting in expanded SLGC-like cells (brackets). Shown are 10-day-old cotyledon abaxial epidermis of wild type (left) and esHDG2-SRDX (right) expressing TMMpro::GUS (top) and EPF2pro::erGFP (bottom). Scale bars: 50 μm. (E) Yeast one-hybrid analysis for activation of lacZ reporter possessing 5′ upstream L1-A-box (white bars), L1-T-box (gray bars) or L1-m-box (black bars) by HDG2, AtML1 or PDF2. SCRM, a known E-box binder (Chinnusamy et al., 2003), was used as a negative control. Bars represent mean values of triplicates. Error bars indicate s.e.m. (F) Yeast one-hybrid analysis for activation of lacZ reporter possessing 5′ upstream HAHR1-A-box (white bars), HAHR1-T-box (gray bars) or HAHR1-m-box (black bars), respectively, by HDG2, AtML1 or PDF2. SCRM was used as a negative control. Bars represent mean value of triplicates. Error bars indicate s.e.m.