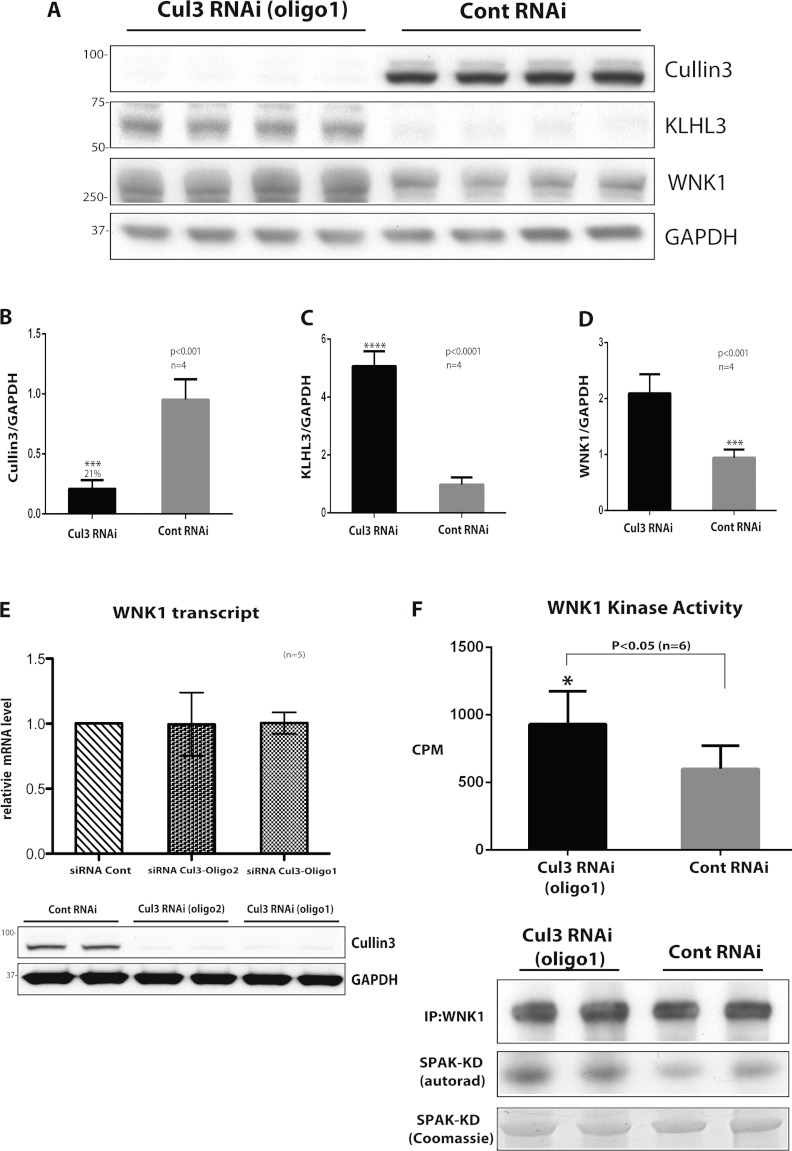
Figure 5. Evidence that CUL3 knockdown enhances WNK1 expression and activity.
HeLa cells were treated with 50 nM scrambled siRNA or CUL3-directed siRNA (probe 1) for 5 days and cells were lysed. (A) Cell lysates were subjected to immunoblotting with the indicated antibody. Each lane contains cell extract from an independent dish of cells from an identical experiment undertaken on the same day. (B–D) Quantitative LI-COR immunoblot analysis was undertaken and the ratio of CUL3 (B), KLHL3 (C) and WNK1 (D) to GAPDH was quantified. Results are means±S.E.M. for two independent samples each assayed in duplicates with P values indicated. (E) Total mRNA was isolated from cells and WNK1 mRNA levels were determined by qRT-PCR. Data were normalized to internal GAPDH and RPL13A controls as described in the Materials and methods section. Results are means±S.D. for three independent samples each assayed in triplicate. (F) WNK1 was immunoprecipitated from 0.25 mg of cell extracts and assayed for its ability to phosphorylate kinase inactive SPAK expressed in E. coli cells. The top panel displays activity in 32P radioactivity incorporated into kinase inactive SPAK (c.p.m.) as mean±S.D. The lower panels display representative immunoblot analysis of WNK1, autorad of phosphorylated SPAK and Coomassie Blue staining of kinase-inactive SPAK employed in the kinase assay. Similar findings were made with an independent CUL3-directed siRNA (probe 2); see data in Supplementary Figure S1 (at http://www.biochemj.org/bj/451/bj4510111add.htm). Cont, control; KD, kinase-dead; IP, immunoprecipitation; RNAi, RNA interference.