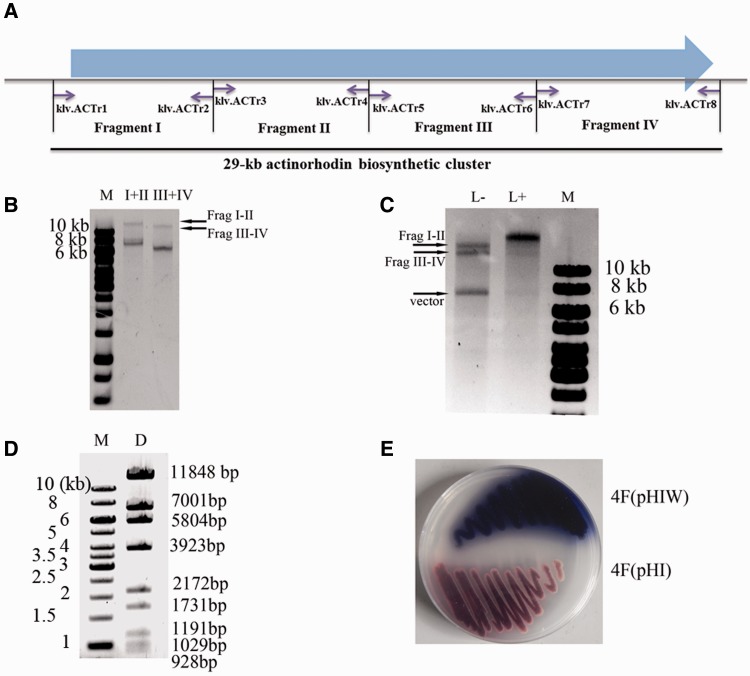
Figure 3.
In vitro seamless assembly of the whole actinorhodin biosynthetic cluster from multiple restriction fragments. (A) The schematic diagram of the PCR amplicons. The 29-kb actinorhodin biosynthetic cluster was divided into four fragments, which were then PCR amplified with primers specified (Supplementary Table S1). (B) Errorless demethylated fragments I (7420 bp), II (8171 bp), III (6410 bp) and IV (6686 bp) were released from pBluescript II KS by XbaI digestion, which were then ligated with a designed adaptor klf.ML2 (Figure 1B and Supplementary Table S1) and digested with MspJI as described in the ‘Materials and Methods’ section. The MspJI-treated fragments were used for ligation with T4 DNA ligase at 16°C for 2 h. (C) Fragments I–II and III–IV were ligated with pHI with (‘L+’) or without (‘L−’) the addition of T4 DNA ligase. The synthesized DNA could be viewed in the ‘L+’ lane together with the disappearance of the substrate fragments. The ligation reaction was performed at temperature 22°C for 8 h. (D) The BamHI restriction map of the synthesized plasmid pHIW. The theoretical restriction fragments of pHIW contains fragments of 11 848, 7001, 5804, 3923, 2172, 1731, 1191, 1029, 928 and 743 bp in size. The 743-bp fragment was run out of the gel and was not shown. (E) Heterologous expression of the assembled actinorhodin biosynthetic cluster in Streptomyces strain 4F (4F/pHIW), using 4F integrated plasmid pHI (4F/pHI) as a negative control. Strains were cultured on R2YE medium at 30°C for 2 days.