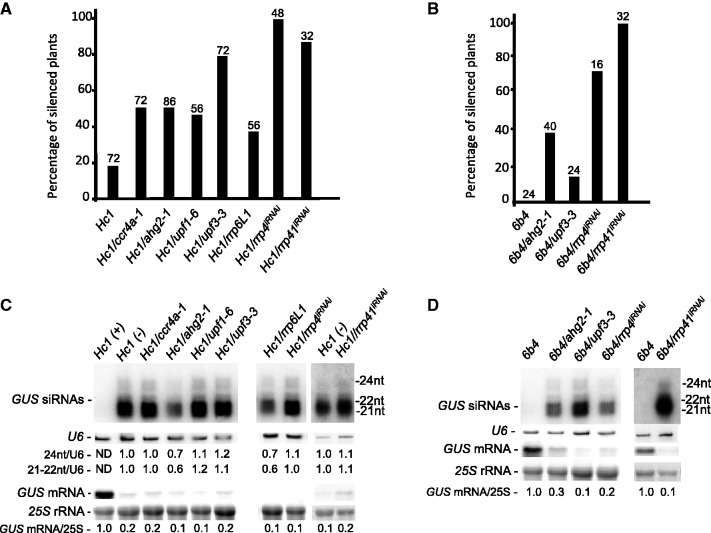
Figure 1.
NMD, deadenylation and exosome mutants enhance transgene S-PTGS. (A and B) The percentage of silenced plants determined by GUS quantitative protein assays in the indicated mutant and control lines. The number of plants analysed is indicated above each bar. (C and D) RNA gel blot analyses of the indicated mutant and control lines. High molecular weight RNA and siRNA gel blots were hybridized with a GUS DNA probe. 25S ribosomal RNA (rRNA) and U6 small nucleolar RNA (snRNA) served as loading controls, respectively. Hc1 plants that were expressing (+) and silenced (−) for GUS were analysed. The position of GUS 24, 22 and 21 nt siRNAs is noted. Normalized values of GUS mRNA to 25S rRNA (with either Hc1 (+) or 6b4 levels set at 1.0) and GUS 24 nt and GUS 21–22 nt siRNA to U6 snRNA [with Hc1 (−) levels set at 1.0] are indicated. ND = non-detectable.