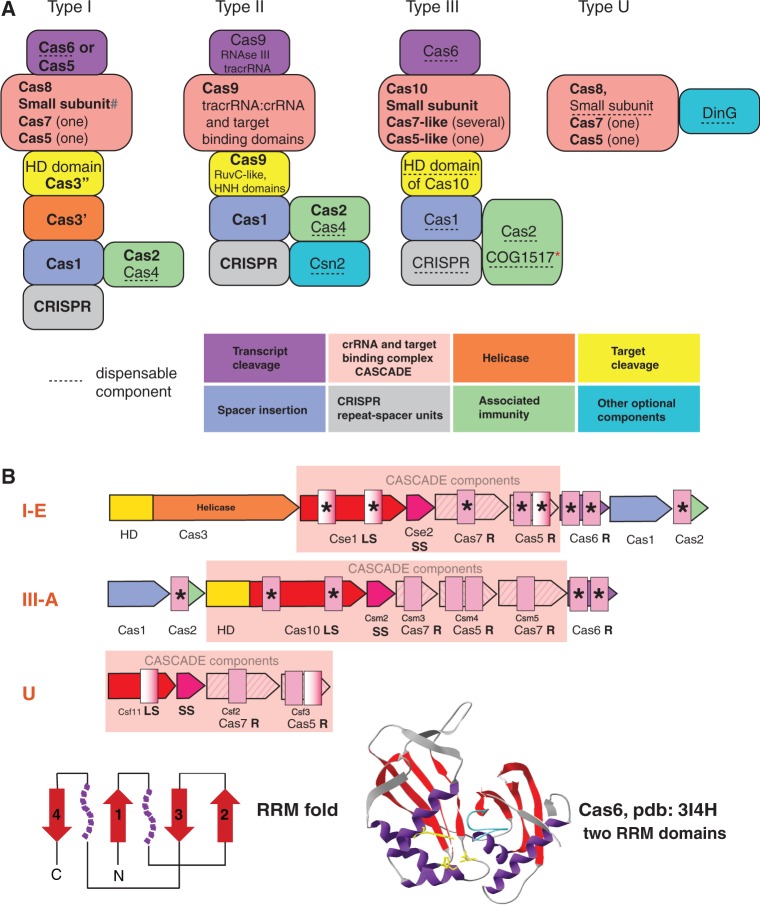
Figure 4.
General principles of the structure and organization of four CRISPR-Cas types. (A) The building blocks of four distinct CRISPR-Cas system types. The cas genes and domain description for each building block are given. Gene names follow the current nomenclature and classification (18). The symbol ‘#’ indicates the putative small subunit that appears to be fused to the large subunit in several Type I subtypes (77). Asterisk indicates that those COG1517 family proteins that contain a third effector (toxin) domain are implicated in immunity-dormancy/suicide coupling. (B) RRM domain-containing proteins in CRISPR-Cas systems. General organization of operons is shown by arrows with size roughly proportional to the size of respective gene. Homologous genes are shown by the arrows of same colour or hashing. Colour coding is the same as in the (A). Gene and family names are taken from (18,77). Additional designations: LS, large subunit; SS, small subunit; R, RAMPs. RRM domains are shown by pink rectangles, with semitransparent rectangles indicating deteriorated RRM fold. The protein representing families with RRM domains for which structures have been solved are denoted by asterisks. A topology diagram of the RRM fold is shown in the bottom left: beta strands are shown by red arrows; the purple shapes each denotes a single alpha helix in the typical RRM fold that, however, are replaced by more complex secondary structure arrangements in some variants including RAMPs. The structure of Cas6, the typical RAMP superfamily protein with two RRM domains, is shown in the bottom right. The colours of the core RRM elements are the same as in the topology diagram; in addition, the glycine-rich loop, the signature feature of the RAMP superfamily proteins, is shown in blue; amino acids involved in catalysis are rendered in yellow.