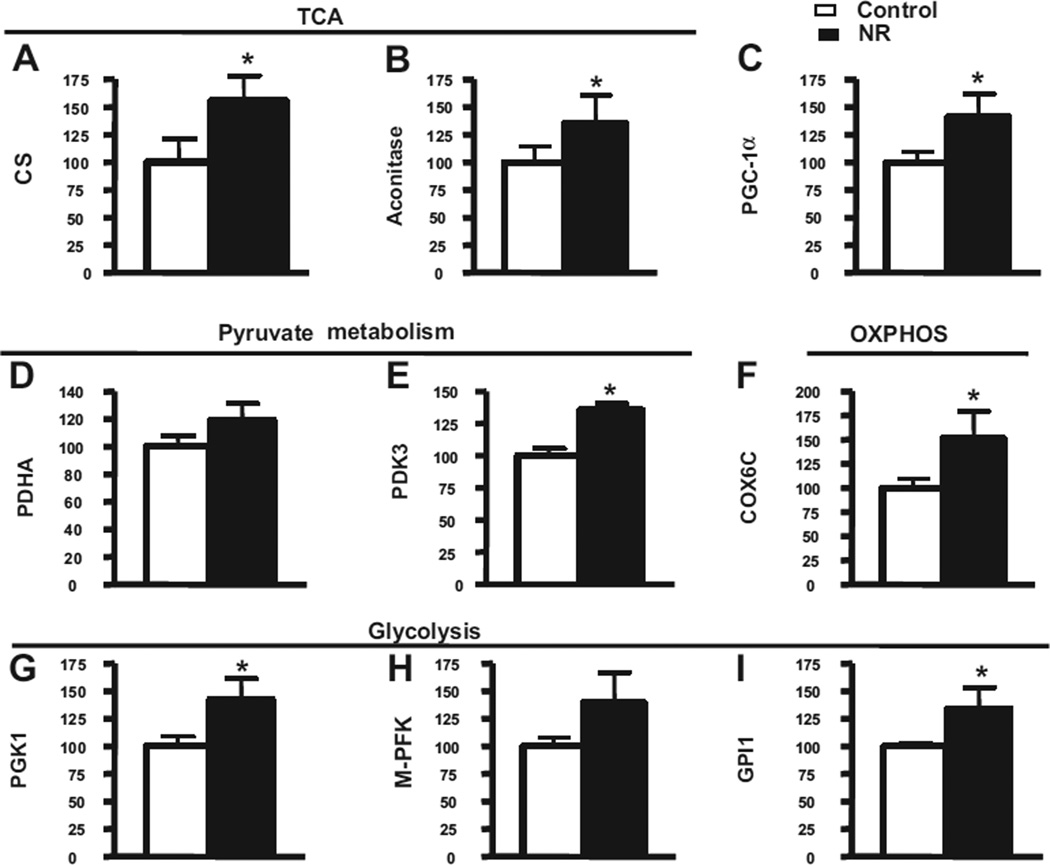
Fig. 7.
Energy metabolism gene products are induced in response to short-term NR treatment in the brains of Tg2576 mice. In this study, 5-month-old Tg2576 mice were treated with 250 mg/kg/day of NR for approximately 3 months, and total RNA from the cerebral cortex was extracted using RNeasy Mini Kit (Qiagen) 24 hours after behavioral testing. Complementary DNA was synthesized using Superscript III First-Strand Synthesis SuperMix for quantitative RT-PCR (Invitrogen) with 1 µg of total RNA. Quantitative RT-PCR was performed using Maxima SYBR Green Master Mix (Fermentas). Gene targets examined here included (A) CS, (B) aconitase, (C) PGC-1α, (D) PDHA, (E) PDK3, (F) COX6C, (G) PGK1, (H) M-PFK, and (I) GPI1. Fold changes between groups were calculated using the ΔCt method. Statistical analysis was done in GraphPad Prism 4. Abbreviations: COX6C, cytochrome c subunit Vic; CS, citrate synthase; GPI1, glucose phosphate isomerase 1; M-PFK, muscle phosphofructokinase; NR, nicotinamide riboside; OXPHOS, oxidative phosphorylation system; PCR, polymerase chain reaction; PDHA, pyruvate dehydrogenase; PDK3, pyruvate dehydrogenase kinase 3; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α; PGK1, phosphoglycerate kinase; RT, reverse transcription; TCA, tricarboxylic acid cycle.