Fig. 5.
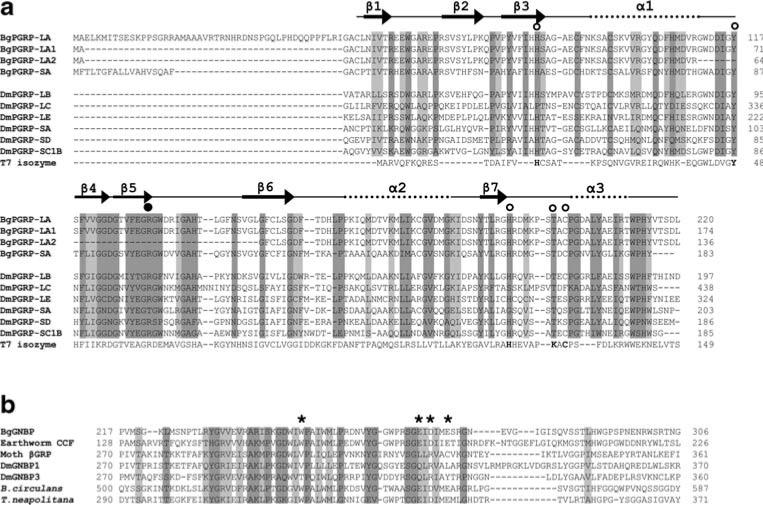
a Structure-based sequence alignment of PGRPs guided by the crystal structures of DmPGRP-LB, -SA, and -LE (Kim et al. 2003; Reiser et al. 2004; Lim et al. 2006). The open circles show the five conserved amino acids, and the filled circle indicates the Arg residue critical for the discrimination of DAP-type PGN. The dark and light shaded boxes show regions of sequence identity and of conserved replacements (BLOSUM62 matrix) of greater than 80%, respectively, in the sequences aligned. The five conserved residues critical for enzymatic activity in T7 isozyme are in bold. The numbers on the right side indicate the position from the start codon in each aa sequence. Dm Drosophila melanogaster. The GenBank accession numbers: DmPGRP-LB (Q9VGN3); DmPGRP-LC (Q9GNK5); DmPGRP-LE (Q9VXN9); DmPGRP-SA (Q9VYX7); DmPGRP-SD (Q9VS97); DmPGRP-SC1B (Q95SQ9); bacteriophage T7 isozyme (P00806). b Alignment of the glucanase-like region of the representative GNBPs. The dark and light shaded boxes show regions of sequence identity and of conserved replacements (BLOSUM62 matrix) of greater than 85% (6/7), respectively. Asterisks indicate the residues required for glucanase activity, as assessed from bacterial studies. The numbers on the both sides indicate the position of amino acids related to the start codon of the aa sequence. Bacillius circulans β-1,3-glucanase A1 (P23903), Thermotoga neapolitana β-glucanase (Z47974), Moth (M. sexta) βGRP (AAF44011), DmGNBP1 (AAF33849), DmGNBP3 (AAF33851)