Abstract
Purpose
Since the identification of TMPRSS2/ERG rearrangement as the most common fusion event in prostate cancer, various methods have been developed to detect this rearrangement and to study its prognostic significance. We hereby report a novel 4-color fluorescence in situ hybridization (FISH) assay that not only detects the typical TMPRSS2:ERG fusion but also alternative rearrangements of either the TMPRSS2 or ERG gene.
Experimental design
We validated this assay on fresh, frozen, or formalin-fixed paraffin-embedded prostate cancer specimens including cell lines, primary prostate cancer, xenograft tissues derived from metastatic prostate cancer, and metastatic tissues from castration-resistant prostate cancer (CRPC) patients.
Results
When compared with RT-PCR or Gen-Probe method as the technical reference, the 4-color FISH assay demonstrated an analytical sensitivity of 94.5% (95% Confidence Interval [CI] 0.80-0.99) and specificity of 100% (95% CI 0.89-1.00) for detecting TMPRSS2:ERG fusion. TMPRSS2:ERG fusion was detected at 41% and 43% in primary prostate cancer (n = 59) and CRPC tumors (n = 82), respectively. Alternative rearrangements other than the typical TMPRSS2:ERG fusion were confirmed by karyotype analysis and shown present in 7% primary cancer and 13% CRPC tumors. Successful karyotype analysis is reported for the first time on four of the xenograft samples, complementing the FISH results.
Conclusions
This 4-color FISH assay provides sensitive detection of TMPRSS2 and ERG gene rearrangements in prostate cancer.
Keywords: Prostate cancer, TMPRSS2, ERG, FISH
Introduction
The chromosomal rearrangements involving androgen-regulated TMPRSS2 and ETS transcription factors present great potential as prognostic markers for human prostate cancer due to their biological relevance, prevalence among prostate cancer patients, specificity to the disease, and underlying molecular mechanisms influencing cellular phenotypes (1). Identification of these rearrangements may allow for stratification of prostate cancers into subtypes that respond to specific therapeutics (2). The most common type of rearrangement involves the fusion of TMPRSS2 to oncogene ERG due to the deletion of intervening genomic sequences on chromosome 21 or insertion of this sequence to separate location(s) of the genome (3, 4), which leads to high levels of ERG expression. TMPRSS2:ERG fusion demonstrates high prevalence and specificity in prostate cancer, such that it has been detected in nearly half of the prostate cancer patients but none of the benign prostate tissues (5-9).
Despite the great interest in clinical significance of the TMPRSS2:ERG fusion, studies on its relevance to disease outcome have yielded discordant conclusions. The inconsistency can be attributed to multiple factors such as the different patient cohort, the determination of study end point, and the technique used to detect TMPRSS2/ERG rearrangement (7). Among the technologies for the detection of gene rearrangements, fluorescence in situ hybridization (FISH) remains the most robust method to date in the analysis of archived clinical samples that are formalin-fixed and paraffin-embedded (FFPE). Of note, the recent development of anti-ERG monoclonal antibodies has further enhanced the detection of ERG overexpression in ERG-rearranged tissues, particularly in FFPE samples (10, 11). In FISH characterized cohorts, these antibodies have shown remarkable correlation between ERG rearrangement and protein expression (11).
The majority of studies that investigated TMPRSS2/ERG rearrangement with FISH used dual-color ERG break-apart probes (12-16), which can identify the rearrangement or deletion of ERG gene but does not inform whether the rearranged ERG is fused with any 5’-partner. A few other studies utilized the tri-color fusion probes (6, 17-19), the application of which is usually challenged by possible overlap in excitation and/or emission wavelengths among different fluorochromes. The largest published study to date utilized a tri-color FISH assay by combining the red/green break-apart probe for TMPRSS2 with an orange-labeled fusion probe for 3’ERG (18).
Here we report a novel 4-color FISH assay that overcomes common technical challenges and allows the detection of either TMPRSS2 or ERG rearrangements regardless of the partner gene, a key advantage. The assay was successfully applied to frozen as well as FFPE prostate cancer samples, including primary cancer and castration-resistant (CRPC) metastatic tumors from patients and xenograft models with high sensitivity and specificity.
Materials and Methods
Sample source: cell lines, xenografts and patient tissues
The study was approved by the Institutional Review Boards at Fred Hutchinson Cancer Research Center (FHCRC) and the University of Washington (UW). LNCaP and VCaP were obtained from American Type Culture Collection (ATCC, VA). De-identified archived primary prostate cancer samples were obtained from UW and Virginia Mason Hospital in Seattle. Prostate cancer xenografts were generated and maintained by the Vessella Lab at UW. Characteristic features of the xenografts have been described previously (20). Tissue microarrays (TMA) were generated from FFPE xenograft tissues as previously described (15). Metastatic CRPC tumor samples were collected from autopsies performed within 2 to 4 hours of death from 11 patients at UW under the rapid autopsy program (21). Tumors were obtained from various organ sites, frozen in OCT and stored at -80°C. All tissues were sectioned for H&E staining and verification of histology reviewed by a pathologist.
Metaphase harvest and karyotype analyses of prostate cancer cell lines and xenografts
LNCaP and VCaP cells were cultured in phenol-red free RPMI 1640 Medium and DMEM (Invitrogen, CA), respectively, supplemented with 10% fetal bovine serum (FBS, HyClone, Thermoscientific, MA) and 1% penicillin/streptomycin/L-glutamine at 37°C in a 5% CO 2 humidified incubator. Cells actively growing in logarithmic phase were incubated with colcemid at the final concentration of 0.04 μg/ml for 60 minutes at 37°C to arrest cells in meta phase. Cells were then treated with the pre-warmed hypotonic solution (0.075 M KCl) for 30 minutes before fixation in methanol and glacial acetic acid (2.5:1 ratio). Fixed cells were dropped on glass slides, treated with 0.025% trypsin in 0.9% NaCl, and stained with 1:4 diluted Wright's stain (pH 6.8) for GTW banding. Metaphases were analyzed under a microscope at a magnification of about 1250X for chromosome count and structural integrity. Karyotypes were written according to the ISCN2009 guideline. Metaphase nuclei were harvested from fresh prostate cancer xenografts in a similar way. Briefly, tumor tissues were thoroughly minced, treated with collagenase (2000 units/ml) to obtain single cell suspensions, which were then cultured in DMEM supplemented with 20% FBS and 1% penicillin/streptomycin/L-glutamine. Cells were allowed to grow for approximately 1 week before exposure to colcemid, followed by hypotonic treatment and fixation.
Fluorescent in Situ Hybridization (FISH)
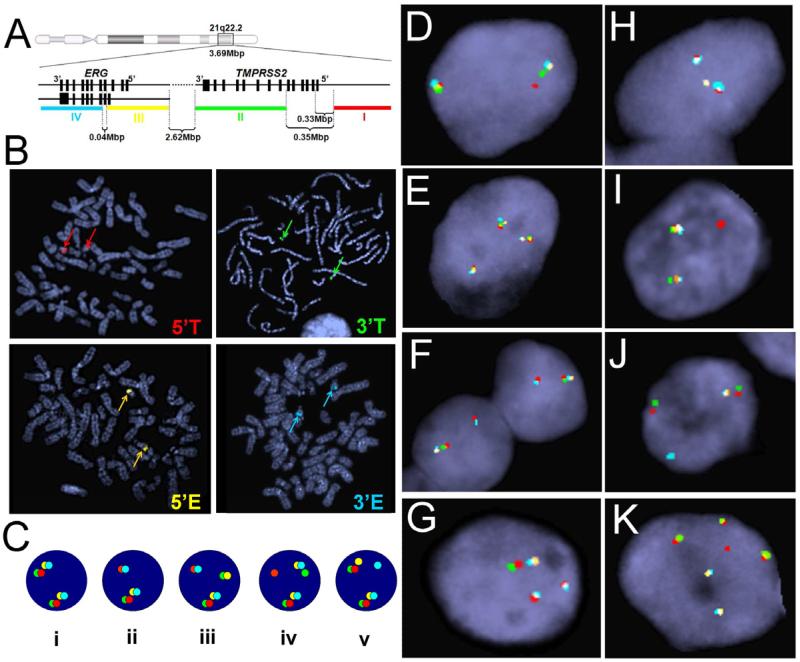
Bacterial artificial chromosome (BAC) clones RP11-35C4, RP11-120C17, RP11-95I21, and RP11-476D17 received as gifts from Dr. Barbara Trask (FHCRC, WA) were used to generate probes targeting 5’-TMPRSS2, 3’-TMPRSS2, 5’-ERG, and 3’-ERG, respectively (Figure 1A). BAC DNA was extracted using procedures recommended by Children's Hospital Oakland Research Institute (CHORI, CA) and labeled with Nick Translation Kit (Abbott Molecular, IL, USA) using SpectrumRed-dUTP (5’-TMPRSS2), SpectrumGreen-dUTP (3’-TMPRSS2), SpectrumGold-dUTP (5’-ERG), and SpectrumAqua-dUTP (3’-ERG) (Abbott Molecular, IL). FISH analysis was performed according to standard procedures as published previously(22). Slides were examined for fusion/rearrangement and enumerated using AxioImager Z1/2 microscopes (Zeiss, Oberkochen, Germany) by two independent scientists. The proper localization of the probes on chromosome 21 was validated by metaphase FISH on normal human peripheral blood lymphocytes (Figure 1B). Sequence specificity of the BAC DNAs was also confirmed by PCR and Sanger sequencing (data not shown). Excluding tissues with unsatisfactory hybridization quality, a total of 59 primary cancer, 26 xenograft samples and 56 metastatic CRPC tumors were fully analyzed by FISH.
Figure 1.
The novel 4-color FISH technique for the detection of rearrangements of TMPRSS2 and/or ERG. (A) The FISH technique involved hybridization of samples with four probes, 5’-TMPRSS2 (red, probe I), 3’-TMPRSS2 (green, probe II), 5’-ERG (gold, probe III), and 3’-ERG (aqua, probe IV) simultaneously. (B) Metaphases prepared from normal human peripheral blood lymphocytes were used to validate proper localization of FISH probes. Red, green, gold, and aqua probes, targeting 5’- TMPRSS2, 3’-TMPRSS2, 5’-ERG, and 3’-ERG, respectively, all located to chromosome 21. Color arrows point to the location of the corresponding FISH signals. (C) Illustration of multiple types of signal patterns detected by 4-color FISH. (D-K) Representative FISH images of tumor cells: (Ci and D) normal; (E) copy number increase (CNI) of normal TMPRSS2 and ERG; (Cii and F) single deletion fusion; (G) dual deletion fusion; (Ciii and H) atypical fusion; (Civ and I) rearrangement of 5’-TMPRSS2; (Cv and J) rearrangement of 3’-ERG; and (K) separation of TMPRSS2 from ERG signals.
Reverse transcription (RT)-PCR and Gen-Probe analyses of TMPRSS2:ERG fusion
Total RNA was extracted from frozen tumor sections using RNeasy Mini Kit (Qiagen, MD). For each sample, 500 ng of RNA was converted to cDNA using SuperScript II Reverse Transcriptase (Life Technologies, CA) with an equal mixture of Random Primers (Life Technologies, CA) and Oligo dT (Integrated DNA Technologies, CA), and then amplified to detect the presence of TMPRSS2:ERG fusion transcript with the following primer set: Fwd 5’-AGC TAA GCA GGA GGC GGA-3’ and Rev 5’-CGA CTG GTC CTC ACT CAC AA-3’.
Two or more 10-micron sections of frozen tissue were placed into an Aptima ® tube (Gen-Probe, CA) containing 3 mL of specimen transport medium. The tubes were heated for 30 minutes at 60C and stored at -80C. The samples were shipped to Gen-Probe. Target Capture RNA isolation, Transcription Mediated Amplification, and TMPRSS2-ERG fusion detection were performed by Gen-Probe as previously described (23). The primers span the junction between TMPRSS2 exon 1 (NM_005656.3) and ERG exon 4 (NM_004449.4), producing an amplification product of 86 nucleotides.
Estimation of the performance characteristics of the 4-color FISH assay
Test sensitivity, specificity, positive/negative predictive value, and the estimated prevalence were calculated using the Clinical Calculator of VassarStats Website for Statistical Computation. Chi-square test was performed using GraphPad Prism.
Results
The novel 4-color FISH assay can detect multiple types of TMPRSS2 and ERG rearrangements
The TMPRSS2/ERG rearrangement was examined by evaluating the number and the relative location of the 4 probes (Figure 1A) that target 5’-TMPRSS2, 3’-TMPRSS2, 5’-ERG, and 3’-ERG on chromosome 21 (Figure 1B) . The normal configuration was represented by the four probes adjacent to or overlapping with each other (Figure 1Ci), which are presented as two 4-color signal clusters in a diploid nucleus (Figure 1D) and three or more signal clusters in an aneuploid nucleus (Figure 1E). Pathologically benign prostate epithelia (n = 10) were used to establish false-positive cutoff values (mean ± 3SD) for various signal patterns on tissue. The typical TMPRSS2:ERG deletion fusion-positive tumors contain at least 50% scored nuclei demonstrating the juxtaposition of 5’-TMPRSS2 (red) and 3’-ERG (aqua) with concurrent loss of the interstitial 3’-TMPRSS2 (green) and 5’-ERG (gold) probes (Table 1 and Figure 1Cii, F, and G). Insertion or translocation, instead of deletion, of the interstitial region to a different genomic location results in an atypical fusion (Figure 1 Ciii and H). Importantly, our unique assay allowed for further categorization of TMPRSS2 and ERG arrangement status beyond merely the presence or absence of gene fusion. When performed on tumor tissue specimens, the complex design of the 4-color FISH technique allowed for the revelation of various TMPRSS2 and ERG signal configurations. In addition to the single deletion fusion and atypical fusion, the subclasses of fusion also include dual or multiple deletion fusion (Figure 1G and Table 1) and complex fusion with a mixture of different fusion and non-fusion alternative rearrangements. Among samples determined as fusion-negative, the assay detected the presence of normal alleles with two copies per nucleus (Figure 1D), copy number increase (CNI) of normal TMPRSS2 and ERG (Figure 1E), and other alternative rearrangements, which include the 5’-TMPRSS2 rearrangement showing individual red signal(s) dissociated from the adjacent green/gold/aqua probe signals (Figure 1Civ and I), and 3’-ERG rearrangement with individual aqua signal(s) separated from the adjacent red/green/gold probe signals (Figure 1Cv and J). In multiple tumors from one CRPC patient (#7), we also observed the separation of TMPRSS2 (red/green) from ERG (gold/aqua) signals (Table 1 and Figure 1K).
Table 1.
Results of TMPRSS2 and ERG rearrangement with the novel 4-color FISH assay on castration resistant prostate cancer samples, including metastasis
| Patient | Tissue |
TMPRSS2:ERG fusion |
Alternative rearrangement | CNI | Normal result | Concordance with RT-PCR/Gen-Probe | ||
|---|---|---|---|---|---|---|---|---|
| Single (del) | Multiple (del) | Atypical | ||||||
| 9 | Adrenal1 | 38% | 56% | Y | ||||
| 9 | Adrenal2 | 38% | 52% | Y | ||||
| 9 | Liver | 64% | 10% | Y | ||||
| 9 | LN1 | 22% | 44% | Y | ||||
| 9 | LN2 | 30% | 54% | Y | ||||
| 9 | LN3 | 34% | 24% | Y | ||||
| 9 | LN4 | 32% | 14% | Y | ||||
| 9 | Lung1 | 32% | 54% | Y | ||||
| 9 | Lung2 | 22% | 66% | Y | ||||
| 9 | Spleen | 40% | 14% | Y | ||||
| 9 | Prostate | 32% | 20% | Y | ||||
| 5 | LN1 | 12% | 80% | Y | ||||
| 5 | LN2 | 100% | Y | |||||
| 5 | LN3 | 20% | 78% | Y | ||||
| 5 | LN4 | 24% | 72% | Y | ||||
| 5 | LN5 | 12% | 88% | Y | ||||
| 5 | Prostate | 84% | 12% | Y | ||||
| 4 | Liver | 92% | Y | |||||
| 4 | LN1 | 96% | N** | |||||
| 4 | Lung1 | 96% | N** | |||||
| 4 | Spleen | 92% | 8% | N** | ||||
| 2 | LN1 | 88% | Y | |||||
| 2 | LN2 | 97% | Y | |||||
| 2 | LN3 | 100% | Y | |||||
| 2 | Lung1 | 79% | Y | |||||
| 2 | Lung2 | 88% | Y | |||||
| 2 | Prostate | 74% | Y | |||||
| 7 | LN1 | 67% | 20% | NA | ||||
| 7 | LN2 | 89% | NA | |||||
| 7 | LN3 | 84% | Y | |||||
| 7 | LN4 | 97% | Y | |||||
| 7 | Prostate | 92% | Y | |||||
| 11 | LN1 | 72% | Y | |||||
| 11 | LN3 | 48% | Y | |||||
| 11 | Prostate 1 | 78% | Y | |||||
| 11 | Prostate 2 | 74% | Y | |||||
| 6 | LN1 | 100% | Y | |||||
| 6 | LN2 | 82% | Y | |||||
| 6 | LN3 | 78% | Y | |||||
| 6 | Peritoneal | 92% | Y | |||||
| 1 | Liver | 12% | Y | |||||
| 1 | LN1 | Normal | Y | |||||
| 1 | LN2 | Normal | Y | |||||
| 1 | Prostate | 24% | Y | |||||
| 3 | Liver | 34% | Y | |||||
| 3 | LN1 | 74% | Y | |||||
| 3 | LN2 | 80% | Y | |||||
| 3 | Lung | 59% | Y | |||||
| 3 | Prostate 1 | Normal | Y | |||||
| 3 | Prostate 2 | Normal | Y | |||||
| 8 | Liver | Normal | Y | |||||
| 8 | LN1 | Normal | Y | |||||
| 8 | LN2 | Normal | Y | |||||
| 8 | LN3 | Normal | Y | |||||
| 8 | Lung | Normal | Y | |||||
| 8 | Prostate | Normal | Y | |||||
| LuCaP49a | Omental fat | 86% | N/Y | |||||
| LuCaP86.2 | Bladder | 62% | 16% | Y | ||||
| LuCaP93a | Prostate | 60% | 36% | Y | ||||
| LuCaP145.1a | Liver | 94% | Y | |||||
| LuCaP145.2a | LN | 14% | 86% | Y | ||||
| LuCaP35 | LN | 100% | Y | |||||
| LuCaP35CR | LN | 100% | Y | |||||
| LuCaP92.1 | Peritoneum | 100% | Y | |||||
| LuCaP58 | LN | 86% | Y | |||||
| LuCaP96 | Prostate | 80% | 26% | Y | ||||
| LuCaP96CR | Prostate | 98% | 38% | Y | ||||
| LuCaP146b | NA | 92% | 28% | NA | ||||
| LuCaP23.12 | Liver | 50% | N | |||||
| LuCaP23.1CR | LN | 60% | N | |||||
| LuCaP69b | NA | 62% | NA | |||||
| LuCaP70 | Liver | 64% | Y | |||||
| LuCaP73 | Prostate | 60% | Y | |||||
| LuCaP105 | Rib | 16% | Y | |||||
| LuCaP115 | LN | 24% | Y | |||||
| LuCaP141 | Prostate | 34% | Y | |||||
| LuCaP77 | Femur | Normal | Y | |||||
| LuCaP78 | Peritoneum | Normal | Y | |||||
| LuCaP81 | LN | Normal | Y | |||||
| LuCaP136 | Acites fluid (cells) | Normal | Y | |||||
| LuCaP147 | Liver | Normal | Y | |||||
| LuCaP153b | NA | Normal | NA | |||||
CR Castration resistant; these xenograft lines used to have the name suffix as “v” or “ai”
LN Lymph node
NA Data not available
Small cell carcinoma samples; discrepancy between FISH and RT-PCR in some (**) was documented previously (Saramäki et al., 2008). Therefore, fusion detected by FISH is considered as true positive.
Xenograft discontinued Blank indicates the corresponding FISH pattern was absent from the sample or below threshold
Validation of the 4-color FISH assay on prostate cancer cell lines
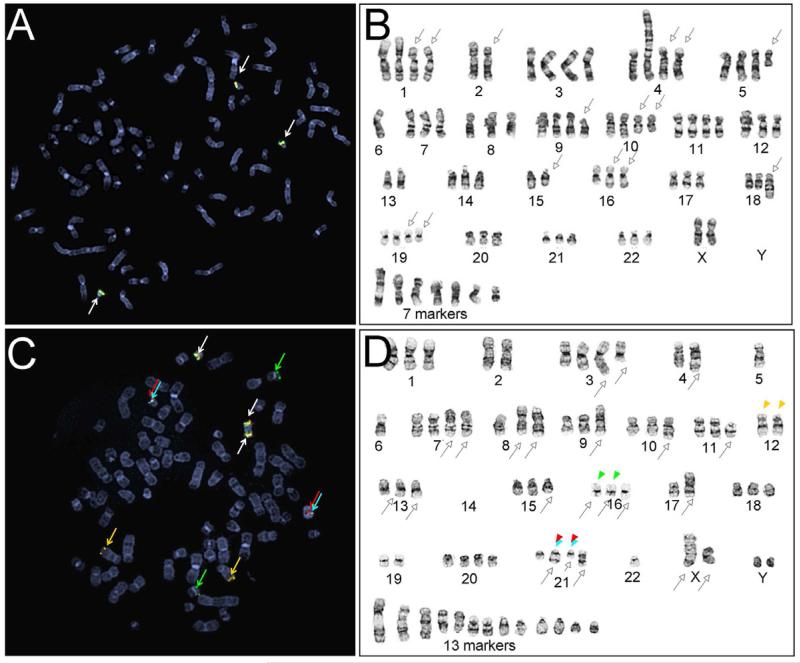
We applied 4-color FISH on prostate cancer cell lines, LNCaP and VCaP, with known TMPRSS2/ERG rearrangement features (5). As expected, LNCaP cells were hypotetraploid without TMPRSS2 or ERG rearrangements displaying 3 to 4 sets of normal TMPRSS2 (red/green) and ERG (gold/ aqua) signals (Figure 2A). Metaphase-FISH further confirmed the presence of normal TMPRSS2 and ERG signals on Chromosome 21 (Figure 2B). VCaP, a fusion-positive cell line, showed a clear TMPRSS2:ERG fusion signal pattern, as well as additional ERG gene rearrangements (Figure 2C). VCaP contained three pairs of the normal TMPRSS2 and ERG, shown as clusters of the four probes, and two fusion signals, demonstrated as juxtaposed 5’-TMPRSS2 (red) and 3’-ERG (aqua) without the interstitial 3’-TMPRSS2 (green) and 5’-ERG (gold) signals. In addition, alternative gene rearrangements were evident, including insertion/translocation of 3’-TMPRSS2 (green) to chromosome 16 and insertion/translocation of 5’-ERG (gold) to chromosome 12 (Figure 2C and 2D).
Figure 2.
Characterization of TMPRSS2 and ERG rearrangement patterns in prostate cancer cell lines determined by the 4-color FISH assay. (A and B) Metaphase FISH and karyogram images from fusion-negative LNCaP; (C and D) Metaphase FISH and karyogram images of fusion-positive VCap. White arrows indicate normal FISH signals, represented as clusters of the 4 probes. Empty arrows with black outline indicate abnormal chromosomes identified during karyotype analysis. Red, green, gold, and aqua arrow or arrow heads highlight positions of FISH signals corresponding to 5’-TMPRSS2, 3’-TMPRSS2, 5’-ERG and 3’-ERG.
Application of the 4-color FISH assay on primary and metastatic prostate cancer samples
Of 59 primary prostate cancer FFPE samples analyzed from 59 patients, 24 (41%) were positive for TMPRSS2:ERG fusion, including 3 (5.1%) with complex rearrangements and 1 (1.7%) with copy number increase (CNI) of the normal TMPRSS2 and ERG loci in addition to fusion; 2 samples (3.3%) showed CNI of TMPRSS2 and ERG without fusion; and 4 samples (7%) showed alternative rearrangements without TMPRSS2:ERG fusion. If we had used the common 2-color ERG break-apart probe, 2 of these 4 samples with 5’-TMPRSS2 rearrangement would have been considered normal.
The 56 metastatic CRPC tumors and 26 xenograft samples (total n = 82) were derived from a total of 32 patients (Table 1 and 2). The 4-color FISH assay detected TMPRSS2:ERG fusion in 35 tumors (43%) from 10 patients (31%) with intra-patient heterogeneity. Among these, dual or multiple fusions (with or without additional normal copies) were present in 20 tumors (24%) from 5 patients (16%). The assay also detected alternative rearrangement without TMPRSS2:ERG fusion in 11 tumors (13%) from 5 patients (16%), which would have been called as normal by 2-color ERG break-apart FISH. Of CRPC samples, 20 tumors (24%) from 11 patients (34%) demonstrated CNI of both TMPRSS2 and ERG without rearrangements. CRPC tissue types include frozen and FFPE sections. When both tissue types were available on the same sample, the results obtained from frozen tissues were consistent with those from FFPE tissues. In particular, the signal patterns observed from the tissue sections were consistent with those seen from intact cultured cells in the 4 xenograft lines described below.
Table 2.
Karyotype results of prostate cancer cell lines and xenografts
| Cell line/xenograft | Karyotype |
|---|---|
| LNCaP | 78~79,XX,-Y,-Y,add(1)(p22)x2,-2,add(2)(p11.2)x2,add(4)(p14),add(4)(q31)x2,del(5)(q14),-6,-6,-6,-7,-8, add(8)(p11.2),add(9)(p13),add(10)(q22)x2,-12,-13,-13,-14,-15,-15, del(15)(q22q24),-16,add(16)(q12)x2,-17,-17,-18,add(18)(q23),del(19)(q13.1q13.1)x2,-21,-22,+6~7mar |
| VCaP | 73,YY,add(X)(q22),del(X)(q22),-1,-2,-2,add(3)(q21),del(3)(q12),-4,-4,add(4)(q21),-5,-5,-5,-6,-6,-6,add(7)(p13),add(7)(p15), -8,add(8)(p11.2),add(8)(p21),-9,add(9)(p24),-10,add(10)(q24),del(11)(p11.2),-12,-12,add(12)(q24.1)x2,-13,add(13)(q32)x2,-14,-14,-14,-14, -15,del(15)(q22),-16,del(16)(q13)x2,i(16)(p10),-17,-17,add(17)(p11.2),-18,-19,-19,add(21)(q22),del(21)(q22),i(21)q10),-22,-22,-22,+13mar |
| LuCaP 23.1 | 76<4n>,XXYY,+X,+X,-1,inv(2)(q13q31)x2,-3,-3,-4,-4,-5,-5,add(5)(p15),add(5)(q11.2),-6,-6,+7,i(8)(q10)x2, -9,-9,-9,-10,-10,-11,-11,-11,-13,-13,add(13)(p11.2)x2,-14,i(14)(q10), -15,-15,-16,-18,add(19)(q13.1),der(19)t(11;19)(q13;q13.3)x2,-22,-22,+7mar |
| LuCaP 35CR | 66<3N>,XX,-Y,-1,add(1)(q32),del(1)(q32),?2,der(2)add(2)(p13)add(2)(q21),+add(3)(q21),add(3)(q21),del(3)(q21q23)x2,add(4)(p14),add(4)(q12), +5x2,add(5)(q13),del(5)(q13q31)x2,add(7)(p13),add(8)(p11.2),del(8)(q13q22)x2,add(9)(q13),add(10)(p11.2),der(10)add(10)(p13)add(10)(q22),-11, -12,add(12)(p11.2),-13x2,14x2,-15,add(15)(p11.2),+16,add(16)(q24),del(16)(q24),-17,-18,add(19)(q13.1),-21x3,-22,add(22)(p11.2),add(22)(q13),+8mar |
| LuCaP 58 | 76~80<4n>,XXY,-Y,add(1)(p13)x2,-2,-2,-2,-3,-3,-4,-5,-5,-5,add(6)(p23)x2,der(7)add(7)(p11.2)del(7)(q11.2q22)x2,
add(8)(p11.2), der(8)t(1 ;8)(p22;p11.2)x2,+10,add(10)(q11.2)x2,-11,add(11)(p11.2),del(12)(p11.2),der(12)t(12;21)(p11.2;q11.2),-13,-13,-15, -15,-16,-16,-17,-17,-18,-18,-19,-21,-21,del(21)(q11.2q22),-22,+9~11mar,3~5dmin |
| LuCaP 96 | 82,XY,-X,-Y,add(1)(p13),der(1)add(1)(p13)add(1)q21),-2,-3,-3,-3,-4,-4,add(4)(p14),del(4)(p14p16),-5,-5,add(5)(q31),-6,-6,add(6)(p25), add(7)(p15),del(7)(q22q32),-8,-8,-8,-9,add(9)(q22),add(9)(q13),add(10)(q22), add(10)(q26),del(11)(q21q23)x3,add(12)(q13),add(13)(q32),-14,-14,-15,-15,-15,-16,-16,-16,-17,add(17)(q21),-18,-18,add(18)(q21),-19, -20,der(20)t(20;21)(p11.2;q22),-21,add(21)(q22),-22,add(22)(q13),+21mar,2dmin |
Concordance between the 4- color FISH and RT-PCR/Gen-Probe Results
Concordant results were observed between the 4-color FISH findings and the TMPRSS2:ERG status defined by either Gen-Probe or RT-PCR analysis (Table 1), focusing on the fusion regardless of other types of rearrangements. The xenografts were generally evaluated using RT-PCR while the metastatic tumors were tested by Gen-Probe although some had both tests performed. Of 77 tumors with both FISH and Gen-Probe/RT-PCR data, 72 showed consistent data between FISH and the reference assay (93.5%), including 40 fusion-negative tumors and 32 tumors with TMPRSS2:ERG fusion. Two samples (2.6%), LuCaP23.12 and LuCaP23.1CR, which are derived from the same tumor, were presented as fusion positive by RT-PCR but not by FISH. Because TMPRSS2:ERG fusion was previously reported in LuCaP23.12 and LuCAP23.1(19, 24), this is considered false-negative result from the 4-color FISH assay. FISH, but not Gen-Probe assay, detected fusion in three tumors (4.0%), all of which were derived from patient #4, an individual with small cell carcinomas of the prostate. Previous studies have demonstrated in small cell carcinomas, even though TMPRSS2:ERG fusion is present at the genomic level, the TMPRSS2:ERG transcript is no longer expressed due to lack of AR or AR signaling that drives transcription expression (19, 25). Therefore, FISH results in patient #4 are considered true positives.
Therefore, using these modified RT-PCR/Gen-Probe results as a technical reference, the 4-color FISH assay has an analytical sensitivity of 94.5% (95% confidence interval [CI] 0.80-0.99) and a specificity of 100% (95% CI 0.89-1.00) for detecting TMPRSS2:ERG fusion. The positive predictive value is ~100% (95% CI 0.88-1.00), and the negative predictive value is 95.2% (95% CI 0.83-0.99). The predicted prevalence of the fusion detected by the 4-color FISH is 48.0% (95% CI 0.37-0.60).
Karyotype analysis of xenograft tissues
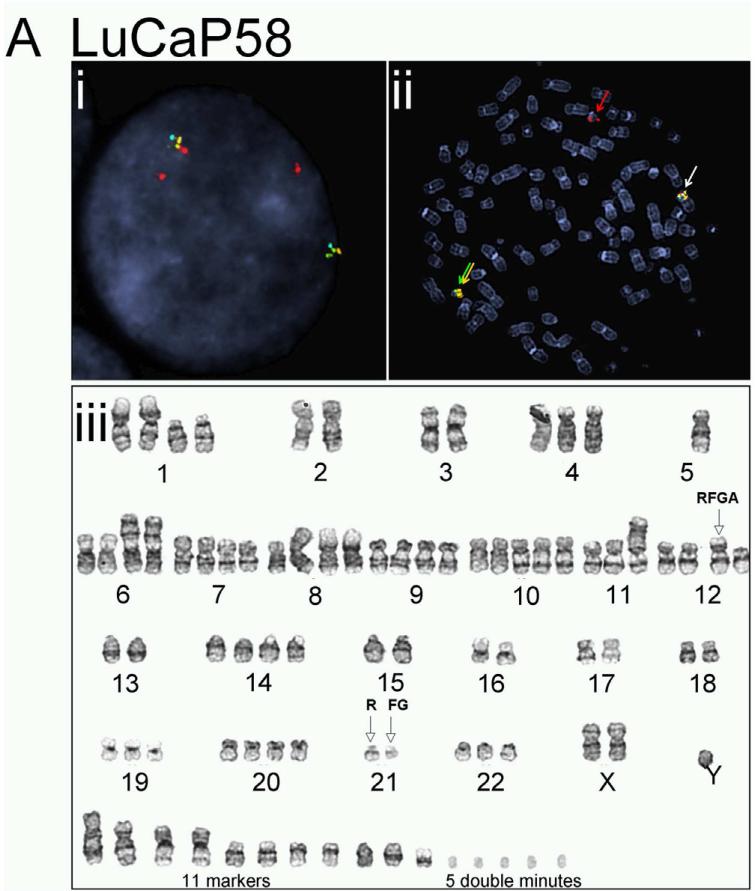
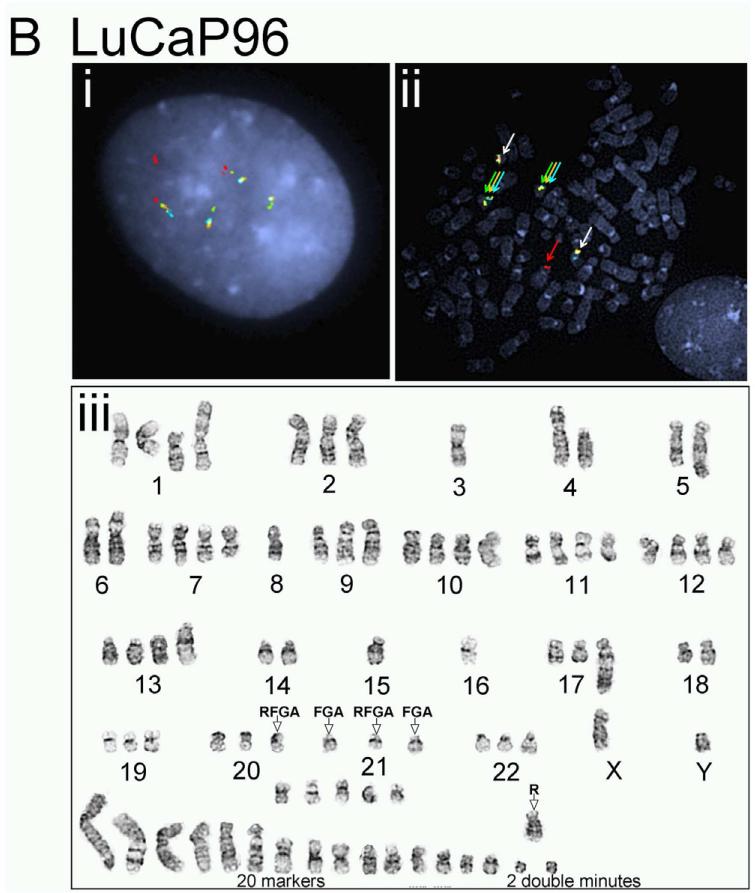
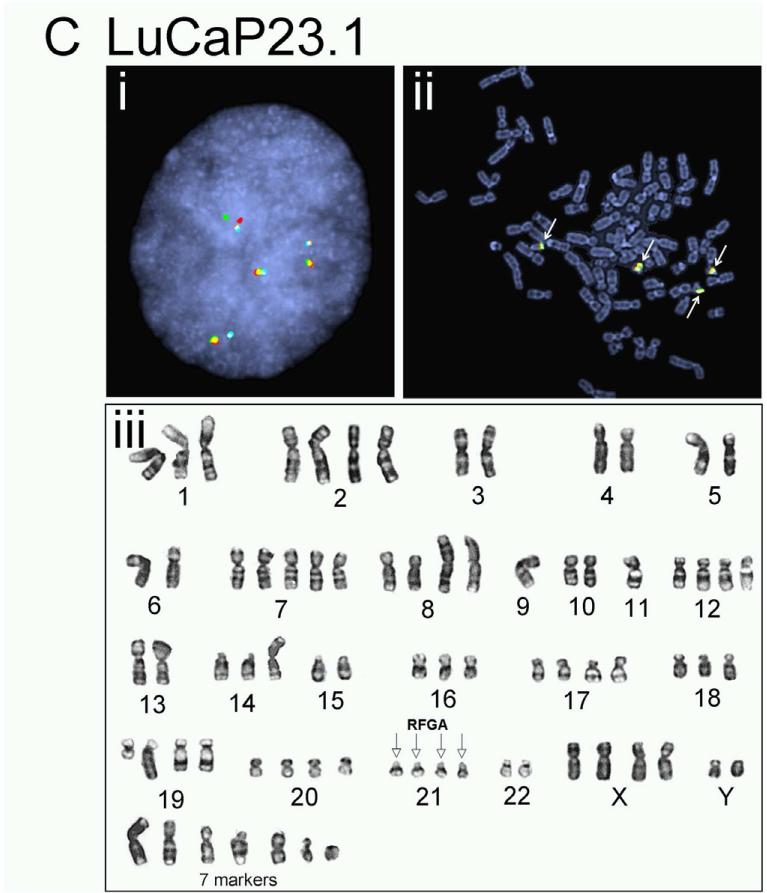
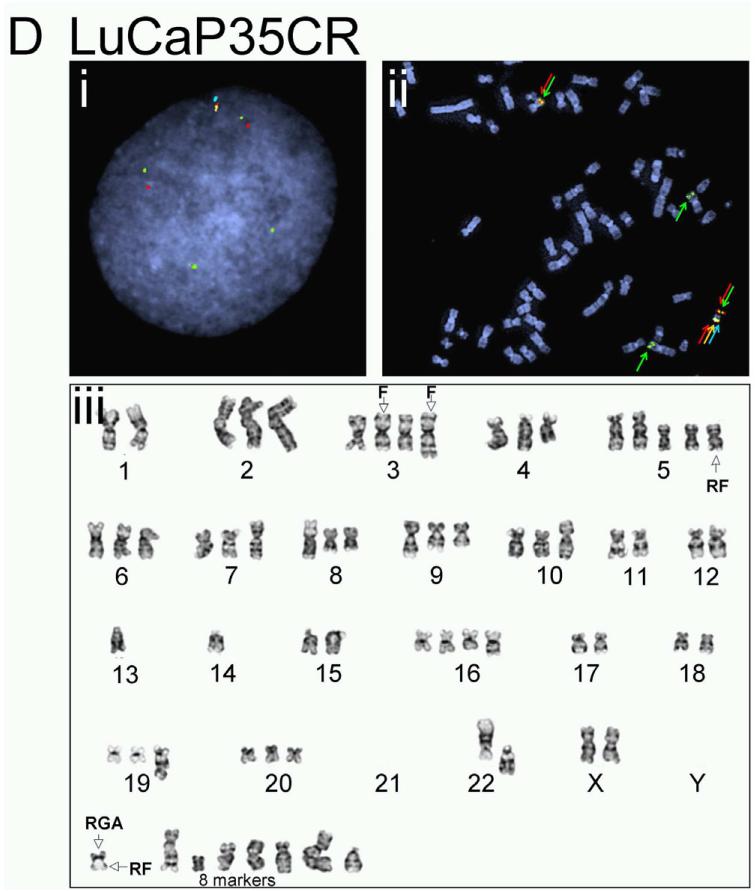
The complicated and potentially novel alternative gene rearrangements called for further characterization using karyotype analysis. Although the TMPRSS2:ERG fusion status has been reported previously for a few LuCaP xenografts(19), the extremely poor in vitro proliferative potential of the prostate cancer xenografts thwarted metaphase analysis for reliable karyotypes. We succeeded in karyotype analyses on four of the LuCaP xenograft tissues, including LuCaP 23.1, 35CR, 58, and 96 (Figure 3 and Table 2). Metaphase and interphase FISH analyses on short-term cultured xenograft cells confirmed alternative TMPRSS2 rearrangement in LuCaP58 (Figure 3A) and LuCaP96 (Figure 3B), atypical fusion in LuCap35CR (Figure 3D), as well as the significant heterogeneity from cell to cell as observed during the FISH analysis on frozen and FFPE tissues (data not shown). Further characterization of the potential partner gene(s) of TMPRSS2 in LuCaP58 and LuCaP96 ruled out two common ETS family members, ETV1 and ETV4, as the rearrangement partner (data not shown).
Figure 3.
Karyotype analysis and corresponding FISH results on prostate cancer xenografts. LuCaP 58 (A), 96 (B), 23.1 (C), and 35CR (D). Representative interphase FISH images obtained from intact nuclei are shown in (i) of each panel; metaphase FISH images are presented in (ii). Karyogram images are in (iii). Red (R), green (F), gold (G), and aqua (A) arrows in FISH images highlight positions of FISH signals corresponding to 5’-TMPRSS2, 3’-TMPRSS2, 5’-ERG and 3’-ERG. Empty arrows with black outline in karyograms highlight the position of indicated signal(s) on a given chromosome.
Discussion
FISH detection of TMPRSS2/ERG rearrangements has the advantage of direct visualization of fusion or other rearrangement events within individual cells in the in situ tissue context. The novel 4-color FISH assay presents a robust tool for the detection of TMPRSS2:ERG fusion that was validated by alternative technologies. In order to identify rearrangements at both fusion gene partners, the 4-color FISH targets both the 5’ and the 3’ regions of TMPRSS2 and ERG, respectively. The spectrally distinct fluorochromes used in our procedure showed little bleed-through across different wavelengths, allowing for a robust assay demonstrating a 94.5% analytical sensitivity and 100% specificity for detecting TMPRSS2:ERG fusion.
The 4-color FISH was successfully performed on frozen samples, FFPE samples, as well as tissue microarrays. The results are highly concordant with RT-PCR and karyotype analysis. In cases where data from frozen and FFPE-sections of the same samples were available, the 4-color FISH showed concordant results regardless of how the samples were preserved. A previous study has demonstrated the TMPRSS2:ERG fusion status in a subpopulation of the xenografts by RT-PCR(19). To demonstrate the technical reliability of the 4-color FISH, we compared its results with RT-PCR/Gen-Probe data obtained from corresponding samples and demonstrated a 93.5% concordant rate. Similar to previous findings, the 4-color FISH detected fusion in samples derived from small cell carcinoma, which was not consistently detectable by RT-PCR or Gen-Probe assay. Interestingly, all false negative samples, those fusion negative by FISH but positive by RT-PCR/Gen-Probe, were derived from the same original CRPC patient. To further understand the discrepancy, we carried out karyotype analysis on LuCaP23.1. Our karyotype analysis showed that LuCaP23.1 contains 4 copies of chromosome 21 per nucleus, each of which presents all four FISH signals. Therefore, although fusion transcript is detectable in the sample, genomic regions corresponding to 5’-TMPRSS2, 3’-TMPRSS2, 5’-ERG, and 3’-ERG all remain on chromosome 21. This is therefore a cryptic rearrangement.
The 4-color FISH assay not only identified TMPRSS2:ERG fusion at a rate similar to previous reports, but also provided researchers the opportunity to further classify fusions into multiple subtypes. Previous findings showed ERG rearrangements in 30-50% of localized prostate cancer (5, 12, 15, 17, 18) and 40-50% among metastatic diseases (13, 18, 24, 26). In the majority of these studies, TMPRSS2/ERG rearrangements were identified via FISH that targeted either ERG only or both TMPRSS2 and ERG but with independent hybridization on separate tissue sections. With this novel 4-color FISH assay reported here, which is capable of detecting rearrangements of TMPRSS2 and ERG simultaneously in a single hybridization, we found rearrangements in 48% primary prostate cancer (including 41% demonstrating actual TMPRSS2:ERG fusion and 7% with alternative rearrangements) and in 57% CRPC metastatic tumors (including 43% TMPRSS2:ERG fusion and 13% alternative rearrangements), consistent with previous findings. Furthermore, the 4-color FISH also identified subtypes of fusion such as single or multiple deletion fusion and single or multiple insertion fusion. For example, the atypical fusion in LuCaP 35CR results from the insertion of the interstitial region between TMPRSS2 and ERG to chromosome 3, which was validated in karyotype analysis. These characteristics enable researchers to compare disease outcomes among patients with different subtypes of fusions in future retrospective and prospective studies (12).
Besides the accurate detection of TMPRSS2:ERG fusion, the 4-color FISH demonstrated unique advantage in identifying multiple types of rearrangements other than TMPRSS2:ERG fusion, such as rearrangement of 5’-TMPRSS2, 3’-TMPRSS3, and 3’-ERG. Karyotype analysis further verified representative alternative rearrangements detected by 4-color FISH. In VCaP, aside from the fusion, the 3’-TMPRSS2 is rearranged to chromosome 16, while the 5’-ERG is at chromosome 12. In LuCaP58 and LuCaP96, 5’-TMPRSS2 is located at a chromosome separate from the 3’-TMPRSS2 and the ERG locus. To the best of our knowledge, our study is the first to present karyotype results of multiple prostate xenografts, which contributes to further understanding of the complex genomic alternations in prostate tumors, especially in the context of whole genome and exome sequencing(3, 27). Additional alternative rearrangements of either TMPRSS2 or ERG uniquely detected by this method may also suggest reflex testing for other less common TMPRSS2 and ERG rearrangements, such as TMPRSS2:ETV fusions, and even allow for identification of potential novel partner genes of TMPRSS2 or ERG. For instance, identification of potential partner gene of TMPRSS2 on chromosome 16 and potential partner gene of ERG on chromosome 12 in the VCaP cell line will be a worthwhile future endeavor.
Variable degrees of difference in TMPRSS2 and ERG rearrangements observed between primary prostate cancer and CRPC in our study is interesting. Even though the frequency of TMPRSS2:ERG fusion is similar in both the primary prostate cancer group and the CRPC group, complex or multiple TMPRSS2:ERG fusion, alternative rearrangement, as well as copy number increase (CNI) without rearrangement, all appeared to be more prevalent in CRPC than in the primary cancer cohort, suggesting a higher level of genomic complexity of CRPC clonal aberrations (Table 3). Multiple metastatic tumors from the same patient nevertheless showed similar TMPRSS2 and ERG rearrangements for the most part. These results are consistent with the hypothesis that TMPRESS:ERG fusion is an early event during cancer development and likely facilitate prostate cancer progression (28). Further rearrangement or duplication of the TMPRSS2:ERG or individual genes, along with additional complex chromosomal translocations may take place as the disease progresses to the more advanced stage, particularly the castration resistant stage.
Table 3.
Differences in the abnormalities detected by the 4-color TMPRSS2/ERG FISH between primary prostate cancer and castration resistant cancer
| Primary tumors |
CRPC tumors |
|||
|---|---|---|---|---|
| N | % | N | % | |
| Total | 59 | 82 | ||
| TMPRSS2:ERG fusion | 24 | 41% | 35 | 43% |
| Single (del) | 13 | 22% | 6 | 7% |
| Multiple (del) | 0 | 0% | 20 | 24% |
| Atypical | 11 | 19% | 9 | 11% |
| Alternative rearrangement | 4 | 7% | 11 | 13% |
| CNI | 2 | 3% | 20 | 24% |
| Normal | 29 | 49% | 16 | 20% |
Only samples successfully hybridized were presented.
Chi-square test results: Chi squared = 115.8, P < 0.0001
In conclusion, the current study demonstrated the development and validation of a novel 4-color FISH assay that can identify fusion as well as multiple types of TMPRSS2 and ERG gene rearrangements with excellent sensitivity and specificity. It provides a powerful tool to study not merely TMPRSS2:ERG fusion, an important specific event in prostate cancer, but also its subtypes and related rearrangements, in the context of prostate cancer pathogenesis and prognosis.
Acknowledgment
We thank Holly Nguyen for maintenance of the xenograft lines and providing specimens, Dr. Colm Morrisey for sectioning the tissue microarray blocks, and Melissa Chiu for PCR and sequencing verification of BAC clones. We also thank Meg Bender and Stacie Thomas for clerical assistance.
This study is supported by PNW SPORE P50 CA097186 and P01 CA 085859 from the National Cancer Institute, and by Richard M Lucas Foundation (for the xenograft generation and clinical specimens).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There is no conflict of interest to disclose.
REFERENCES
- 1.Shah RB, Chinnaiyan AM. The discovery of common recurrent transmembrane protease serine 2 (TMPRSS2)-erythroblastosis virus E26 transforming sequence (ETS) gene fusions in prostate cancer: significance and clinical implications. Adv Anat Pathol. 2009;16:145–153. doi: 10.1097/PAP.0b013e3181a12da7. [DOI] [PubMed] [Google Scholar]
- 2.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, Liu M, Lonigro R, Prensner JR, Tomlins SA, Chinnaiyan AM. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res. 2012;72:1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teixeira MR. Chromosome mechanisms giving rise to the TMPRSS2-ERG fusion oncogene in prostate cancer and HGPIN lesions. Am J Surg Pathol. 2008;32:642–644. doi: 10.1097/PAS.0b013e31815b6056. author reply 644. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Rajput AB, Miller MA, De Luca A, Boyd N, Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME, Cox ME, Huntsman DG. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Pavlovitz B, Tull J, Wang Y, Deng FM, Fuller C. Detection of TMPRSS2 gene deletions and translocations in carcinoma, intraepithelial neoplasia, and normal epithelium of the prostate by direct fluorescence in situ hybridization. Diagn Mol Pathol. 2010;19:151–156. doi: 10.1097/PDM.0b013e3181bb216a. [DOI] [PubMed] [Google Scholar]
- 9.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 10.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, Thangapazham R, Chen Y, McMaster G, Sreenath T, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate cancer and prostatic diseases. 2010;13:228–237. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, Shah RB. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 14.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 15.Esgueva R, Perner S, C JL, Scheble V, Stephan C, Lein M, Fritzsche FR, Dietel M, Kristiansen G, Rubin MA. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23:539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Chae JY, Kwak C, Ku JH, Moon KC. TMPRSS2-ERG gene fusion and clinicopathologic characteristics of Korean prostate cancer patients. Urology. 2010;76:1268, e1267–1213. doi: 10.1016/j.urology.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 17.FitzGerald LM, Agalliu I, Johnson K, Miller MA, Kwon EM, Hurtado-Coll A, Fazli L, Rajput AB, Gleave ME, Cox ME, Ostrander EA, Stanford JL, Huntsman DG. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer. 2008;8:230. doi: 10.1186/1471-2407-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, Eastham JA, Scardino PT, Scher HI, Tickoo SK, Reuter VE, Gerald WL. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 20.Corey EV,R. Xenograft Models of Human Prostate Cancer. In: Chung LWK IW, Simons JW, editors. Contemporary Cancer Research: Prostate Cancer: Biology, Genetics, and the New Therapeutics. I. Humana Press; Totowa: 2007. pp. 3–31. [Google Scholar]
- 21.Morrissey C, True LD, Roudier MP, Coleman IM, Hawley S, Nelson PS, Coleman R, Wang YC, Corey E, Lange PH, Higano CS, Vessella RL. Differential expression of angiogenesis associated genes in prostate cancer bone, liver and lymph node metastases. Clin Exp Metastasis. 2008;25:377–388. doi: 10.1007/s10585-007-9116-4. [DOI] [PubMed] [Google Scholar]
- 22.Ronski K, Sanders M, Burleson JA, Moyo V, Benn P, Fang M. Early growth response gene 1 (EGR1) is deleted in estrogen receptor-negative human breast carcinoma. Cancer. 2005;104:925–930. doi: 10.1002/cncr.21262. [DOI] [PubMed] [Google Scholar]
- 23.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, Brentano S, Mathis J, Pham J, Meyer T, Cass M, Hodge P, Macairan ML, Marks LS, Rittenhouse H. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 24.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 25.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 26.Holcomb IN, Young JM, Coleman IM, Salari K, Grove DI, Hsu L, True LD, Roudier MP, Morrissey CM, Higano CS, Nelson PS, Vessella RL, Trask BJ. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer Res. 2009;69:7793–7802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, White TA, MacKenzie AP, Clegg N, Lee C, Dumpit RF, Coleman I, Ng SB, Salipante SJ, Rieder MJ, Nickerson DA, Corey E, Lange PH, Morrissey C, Vessella RL, Nelson PS, Shendure J. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]