Abstract
Explanations for the genetic variation ubiquitous in natural populations are often classified by the population–genetic processes they emphasize: natural selection or mutation and genetic drift. Here we investigate models that incorporate all three processes in a spatially structured population, using what we call a construction approach, simulating finite populations under selection that are bombarded with a steady stream of novel mutations. As expected, the amount of genetic variation compared to previous models that ignored the stochastic effects of drift was reduced, especially for smaller populations and when spatial structure was most profound. By contrast, however, for higher levels of gene flow and larger population sizes, the amount of genetic variation found after many generations was greater than that in simulations without drift. This increased amount of genetic variation is due to the introduction of slightly deleterious alleles by genetic drift and this process is more efficient when migration load is higher. The incorporation of genetic drift also selects for fitness sets that exhibit allele-frequency equilibria with larger domains of attraction: they are “more stable.” Moreover, the finiteness of populations strongly influences levels of local adaptation, selection strength, and the proportion of allele-frequency vectors that can be distinguished from the neutral expectation.
Keywords: maintenance of genetic variation, selection, drift, spatial heterogeneity, mutation
IN his landmark survey, The Genetic Basis of Evolutionary Change, Richard Lewontin (Lewontin 1974) argued that the failure to understand the processes responsible for maintaining the high levels of genetic variation ubiquitous in natural populations was the central problem in population genetics. In his view, neither neutral nor selective explanations fully dealt with the observations at the time (Lewontin 1974). In the intervening decades, both sorts of explanations have been modified in attempts to resolve Lewontin’s “paradox of variation.” For example, the “nearly-neutral theory” expands the origin neutral theory to include slightly deleterious and advantageous alleles in the standing variation (Ohta and Gillespie 1996).
One problem facing selectionist explanations is that the parameter space that allows the maintenance of polymorphisms by viability selection is vanishingly small for even moderate levels of variation (Lewontin et al. 1978). The power of such explanations, however, has been augmented by the realization that selectively maintained polymorphisms may build up over time, accumulating rare beneficial mutations (Spencer and Marks 1993). By bombarding populations under selection with a steady stream of novel mutations, using what we here call a construction approach, the parameter space that allows selectively maintained polymorphisms can be more easily reached (Spencer and Marks 1988). This approach is analogous to one used in ecological theory by which multispecies communities are constructed by introducing one species at a time to an already stable community (Taylor 1985). Moreover, this construction approach has been applied to more sophisticated models of selection, such as frequency-dependent selection (Trotter and Spencer 2008) and spatially varying selection (Star et al. 2007a). This latter form of selection may be particularly important: spatial structure and its consequent constrained gene flow allow natural populations to adapt to the local conditions that generated the spatial heterogeneity in selection pressures (e.g., Labbe et al. 2005; Hall and Willis 2006). We note, too, that the importance of population structure in maintaining genetic variation continues to be of interest to theorists (e.g., Bürger 2009; Peischl 2010; Bonneuil 2012). Our previous simulation work (Star et al. 2007a,b, 2008) showed that, compared to panmictic populations, structured populations may possess a significantly greater number of selectively maintained alleles. Moreover, as expected from analytic theory (Felsenstein 1976; Hedrick et al. 1976), the levels of variation in these simulations are higher when, relative to the differences in selection strength, gene flow between demes is lower (Star et al. 2007a,b, 2008).
Nevertheless, all of the construction work to date has ignored the stochastic effects of genetic drift, which is likely to be a critical determinant of the levels of polymorphism even for selectionist explanations. First, polymorphism in construction models arises from the accumulation of beneficial mutants, which (everything else being equal) is more likely in larger populations because any reduction in frequency due to genetic drift—and hence possible extinction—is (on average) smaller. Second, mutations—both deleterious and beneficial—arise in direct proportion to population size for a given mutation rate. In smaller populations both these factors, the greater effect of genetic drift and the smaller absolute numbers of mutations, may severely limit the role of environmental heterogeneity in selectively maintaining genetic variation (Leimu and Fischer 2008). In this article we expand the construction approach as applied to spatially varying selection to incorporate these consequences of population size.
Specifically, we investigate the effect of three different population sizes and seven levels of gene flow on the amount of genetic variation maintained by a two-deme construction model incorporating recurrent mutation. We examine the population mean fitnesses that result from this model in order to see how the different population sizes influence the level of adaptation achieved. We also analyze the strength of selection and investigate whether the resulting allele-frequency patterns can be distinguished from those predicted by the neutral theory (Ewens 1972). For each population size, models were run with and without genetic drift to investigate which consequence of population size—genetic drift or mutation number—has a larger influence on the role of spatial heterogeneity in the selective maintenance of genetic polymorphisms.
Model
The selection model used here is based on the standard recurrence equations for constant-viability selection at a single, diploid locus (Hartl and Clark 2007), which was adapted for two equally sized demes. Selection acts locally and this model is therefore one of soft selection. Generations are discrete and the frequency of the ith allele (i = 1, 2, ..., n), Ai, in the dth deme (d ∈{1, 2}), after selection is given by
| (1) |
in which pi,d is the current frequency of Ai in the dth deme, is the current marginal fitness of Ai in the dth deme, wij,d = wji,d is the fitness of the AiAj genotypes in the dth deme, and is the current mean fitness of the dth deme.
Gene flow follows selection, and a proportion (m) of the frequency vector pi,d is divided over both demes, giving the new frequency of Ai in deme d,
| (2) |
where if d = 1 and vice versa.
We implemented this model in a computer simulation (File S1), to investigate the effects of recurrent mutation and population size. Simulations were initiated with a single allele (with a frequency, therefore, of 1.0) and a homozygote fitness of 0.5 in both demes. Then, for a population of total size N with n alleles (n being initially 1), a new mutation, An + 1, was added to a single randomly chosen deme with an initial allele frequency of 1/2N. For these new mutations, new fitnesses (wi(n+1),d for i = 1,..., n + 1 and d = 1, 2) were independently drawn from the uniform distribution on [0, 1]. We do not for a moment suggest that fitnesses of real mutations are so distributed, but this procedure allows us to sample evenly across parameter space. New mutations were introduced to the model using a Poisson process with a mutation rate (μ) of 5 × 10−6/locus. After selection and migration, genetic drift in a population of size N/2 with n alleles was simulated by taking a sequence of n − 1 nested binomial samples of the allele frequencies each generation (see Gentle 2003, p. 198). Allele Ai was considered extinct if .
The model was run with three different total population sizes (N) of 103,104, and 105 and these populations were divided into two equally sized demes (each with respective sizes of 5 × 102, 5 × 103, and 5 × 104, therefore). Thus, when N = 103, for example, one mutation arose every 100 generations, on average. Simulations were run for each value of N and seven different levels of gene flow (m ∈ {0, 0.01, 0.05, 0.1, 0.2, 0.5, 1.0}) for 104 generations with 103 replicates for each combination of m and N. To compare these results to an unstructured population, 103 replicates were also run using a single-deme (panmictic) model, for each value of N. Because we used a fixed mutation rate, μ, in all our simulations, the total number of mutations encountered after 104 generations is directly proportional to N. To differentiate the consequences of genetic drift from the effects of encountering different numbers of mutations, a third set of simulations were run without drift (i.e., a fully deterministic selection model with the same μ and allele-frequency extinction settings).
After 104 generations, the number of alleles (n) present, the frequency vectors, and the fitness sets, which consist of all pairwise viabilities, were recorded for further analysis for all single and two-deme populations (either with or without drift). To compare levels of fitness between single and two-deme populations, we calculated mean fitness by averaging mean fitness per deme over both demes for the two-deme populations. To quantify the strength of selection in our simulated populations for which n > 1, we first defined the relative fitness of genotype AiAj in deme d as ; the corresponding selection coefficient, and has domain [0, 1] (Stoffels and Spencer 2008). We then took the average of the n(n + 1)/2 sij,d values as our measure of the strength of selection within each deme, sd, and these two values were averaged to get an overall mean measure, s.
The recorded fitness sets, after 10,000 generations, were classified according to their ability to deterministically maintain equilibrium conditions. One characteristic of polymorphisms in a spatial, two-deme model with spatially varying selection pressures, is that some fitness sets can generate equilibria that are locally stable rather than globally stable; i.e., only a subset of possible initial allele-frequency vectors converge toward such equilibrium. The occurrence of locally stable equilibria was investigated by reiterating each recorded fitness set with 250 random initial allele-frequency vectors. These initial allele-frequency vectors were generated using the “broken stick method” (Holst 1980) and each of the 250 evaluations were deterministically iterated, without mutation until equilibrium, or until any allele, Ai, became extinct at . Thus, we can define three types of fitness sets as in (Star et al. 2007a): all initial allele-frequency vectors iterated to a fully polymorphic equilibrium for type I fitness sets; some, but not all, initial allele-frequency vectors did so for type II fitness sets; and no vectors did for type III fitness sets. Type III fitness sets may occur because the iterations were stopped after an arbitrary time and equilibrium conditions may not have been reached (Marks and Spencer 1991). Therefore, deleterious alleles may be present in the recorded fitness sets after 10,000 generations. We characterized the ability to deterministically maintain variation under this model by calculating the proportion of types of fitness sets for each combination of m and N.
Finally, we used the Ewens–Watterson test (Ewens 1972; Watterson 1977, 1978) to investigate whether the random effects of genetic drift influence the proportion of recorded allele-frequency vectors (for which n > 1) that can be distinguished from those expected under the neutral theory. Neutrality was examined using two measurements; one measurement used the allele-frequency vector, pd = (p1,d, p2,d, …, pn,d), taken from one of the demes, and the other used the pooled frequency vectors from both demes. From both allele-frequency vectors (i.e., an unpooled and a pooled frequency vector) per recorded simulation, 200 sample frequency vectors were taken, each with a sample size of 200 genes, following a method described by Marks and Spencer (1991) and Spencer and Marks (1992). For each of these sample frequency vectors, the sample homozygosity was calculated and compared to the lower and upper critical points of Ewens sampling distributions (Ewens 1972; Watterson 1977, 1978). Thus, each single allele-frequency vector is tested 200 times. If significantly >5% of the sample frequency vectors for a particular allele-frequency vector are in either critical region, that vector is detectably different from a vector expected under the neutral hypothesis. This detection occurs when >9% of the sample frequencies are rejected (this value coming from the binomial distribution with a sample size of 200 and a probability of success of 0.05). In essence, allele-frequency vectors are detected as nonneutral if their frequency distribution is considered too even or too skewed to be generated by the neutral hypothesis.
Results and Analysis
Numbers of alleles
The different population sizes (N) substantially influence the recorded number of alleles (n) after 104 generations of mutation and selection, regardless of the presence or absence of genetic drift and irrespective of whether the model has one or two demes (Figure 1A; see Supporting Information, Figure S1 for some typical examples of individual simulations). This finding is not surprising considering the large difference in number of mutations encountered during the simulations for the different population sizes. We are more interested in the relative effects of genetic drift in the different population sizes on the generated levels of genetic variation compared to the non-drift equivalent models of the “same size” (i.e., those that, on average, are bombarded with the same number of mutations).
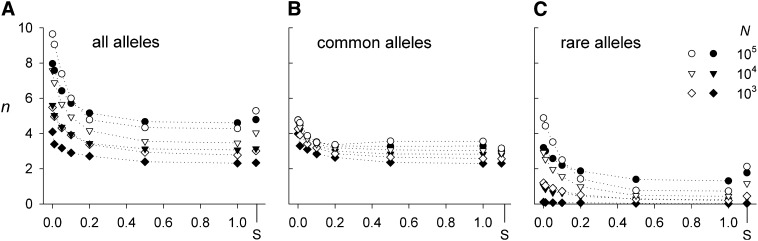
Figure 1.
The number of alleles (n) maintained after 10,000 generations as a function of gene flow (m) for three different total population sizes (N). Results are shown for (A) all alleles, (B) common alleles (i.e., with frequencies >5%), and (C) rare alleles (i.e., with frequencies <5%). Standard errors are small (<0.07) and omitted for clarity. The solid symbols indicate the results for populations with genetic drift whereas the open symbols indicate those without. The letter S on the x-axis indicates the results for single-deme populations.
Genetic drift usually reduces the total amount of genetic variation and this effect is comparably stronger for the lower levels of gene flow (m) when spatial structure becomes more pronounced. Interestingly, for N = 105 and m > 0.1, the two deme simulations with genetic drift actually finish with higher levels of polymorphism compared to their non-drift equivalents. This counterintuitive result, however, is not present when only common alleles (i.e., with frequencies of > 5%) are considered (Figure 1B) and is driven by an increased accumulation of rare alleles (Figure 1C). In contrast, for the smallest populations (N = 103), the presence of rare alleles is limited, especially when these populations experience genetic drift. Overall, the process of mutation accumulation results in a relatively larger proportion of rare alleles (i.e., with allelic frequencies <5%) with increasing population size, in particular for low levels of gene flow.
Fitness
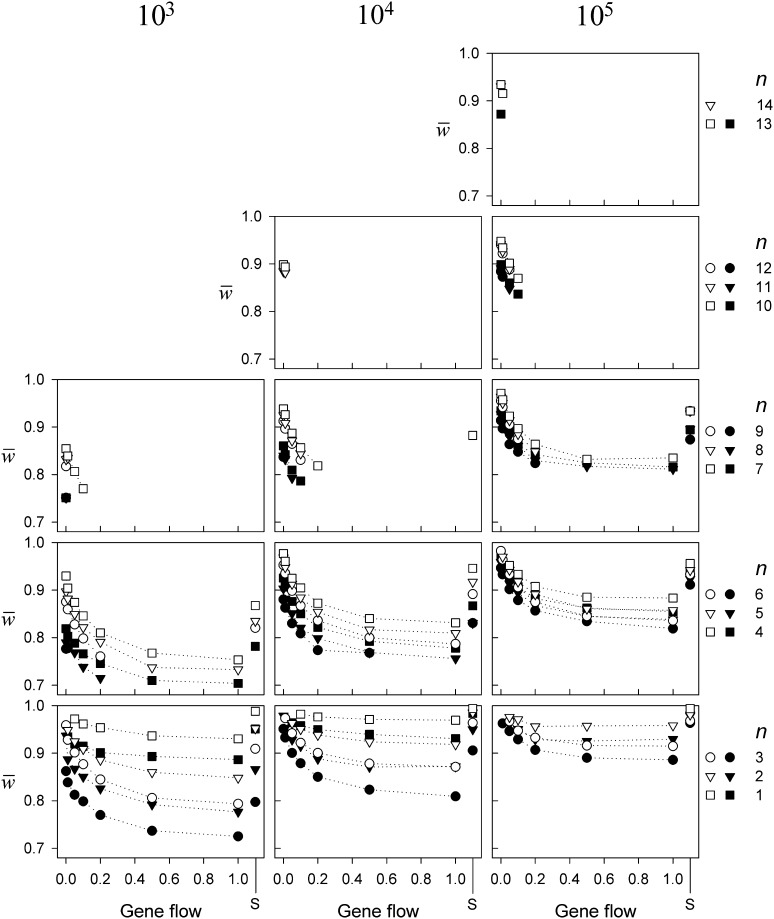
The patterns of recorded mean fitness after 104 generations are complex and influenced by population sizes (N), gene flow (m), number of alleles (n), drift, and whether the model has one or two demes (Figure 2). Nevertheless, some more pronounced patterns are easily recognized: first is the large effect of n on , which decreases with increasing n. Second, decreases with greater gene flow (m), an effect more pronounced at higher values of n (i.e., n > 3). Third, increases with increasing N, a result reflecting the higher numbers, fitter mutants, and their greater survival probabilities in the larger populations during the 104 generations. Last, and most importantly, genetic drift always lowers the level of . The reduction in can be caused by two processes: (i), drift may prevent the invasion of new beneficial mutants or cause successful, well-adapted mutants to go extinct, and (ii) drift may allow slightly deleterious alleles to invade and potentially reach high frequencies. Either process may lower the level of fitness compared to the non-drift equivalent models. While this effect of drift is relatively stronger for lower N and lower levels of m, it is nevertheless not strong enough to prevent local adaptation; regardless of N, the highest levels of are found at low levels of m, indicating substantial levels of local adaptation.
Figure 2.
The level of mean fitness after 10,000 generations as a function of gene flow for three different total population sizes (N). Standard errors are small (<0.01) and omitted for clarity. The solid symbols indicate the results for populations with random drift whereas the open symbols indicate those without. The letter S on the x-axis indicates the results for single-deme populations. Only combinations of N, m, and n are shown for which at least 10 replicates were found.
Selection strength
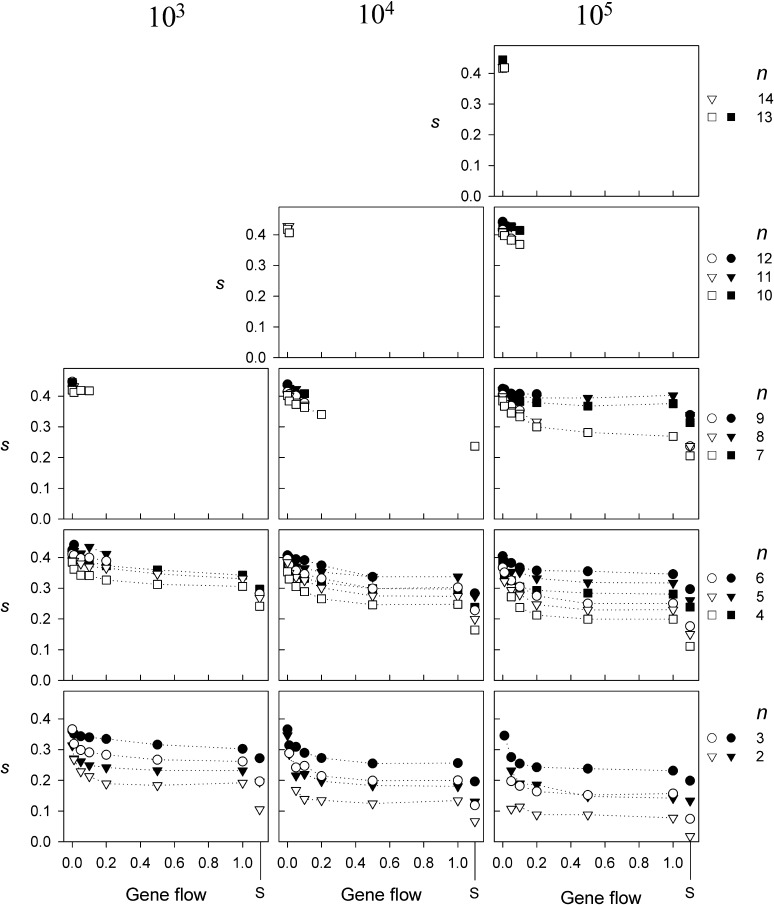
Measures of pairwise selection strength (s) between the genotypes indicate how much variation exists within the fitness sets of the single or two-deme models, normalized by the highest genotypic fitness in a particular fitness set. As with levels of fitness, levels of s are influenced by population sizes (N), gene flow (m), number of alleles (n), genetic drift, and whether the model has one or two demes (Figure 3). Obviously, the two-deme models maintain variation with higher levels of s compared to single-deme models, a feature mainly due to local adaptation and local selection. Most striking, however, are the large differences in s between models with and without drift for the highest N and high levels of m. The higher levels of s can be explained by the presence of a substantial number of slightly deleterious alleles in these populations due to the random effects of genetic drift. Slightly deleterious alleles will have fitness values that are relatively low compared to the highest fitness in that deme and therefore increase the selection strength (s) in the fitness sets, regardless of the allelic frequency with which they occur. While these mutations are constantly selected against, a higher number of novel mutations arising in the larger populations ensures a more constant supply of new maladapted mutants compared to the smaller population sizes. Moreover, at the levels of gene flow (m > 0.1) where the effect of drift on s is most profound, the levels of fitness are relatively low and therefore these mutants are selected against less effectively.
Figure 3.
The level of selection strength (s) for all genotypes after 10,000 generations as a function of gene flow for three different total population sizes (N) and n > 1. Standard errors are small (<0.012) and omitted for clarity. The solid symbols indicate the results for populations with random drift whereas the open symbols indicate those without. The letter S on the x-axis indicates the results for single-deme populations. Only combinations of N, m, and n are shown for which least 10 replicates were found.
These results show that selection models with spatial heterogeneity in selection pressures are more easily invaded by slightly deleterious mutants when levels of migration load are high. This increased potential to be invaded by deleterious alleles is also reflected in the higher numbers of alleles maintained as mentioned above. Nevertheless, the deleterious alleles do not attain high frequencies, since previous results show that the level of mean fitness is not severely decreased relative to the same model without genetic drift for these population sizes and higher levels of gene flow. The rareness of these alleles is also confirmed by calculating the levels of s using only the fitnesses of genotypes comprising common alleles (i.e., with allelic frequencies of >5%); most differences in s between models with or without drift then disappear (Figure S2).
Stability of equilibria
The stability of the recorded polymorphisms was investigated by calculating the proportion of type I, II, or III fitness sets for the different population sizes (N), levels of gene flow (m), and whether genetic drift is present or absent. Genetic drift has a qualitatively different effect on the proportion of equilibrium types for the largest population size (N = 105) compared to the smaller population sizes (N = 103, 104); for these smaller population sizes, genetic drift mostly reduces the relative proportion of type II fitness sets (except when m = 0), which maintain only their selective polymorphism within a neighborhood of their equilibrium (Table 1). Thus, once genetic drift has moved the allelic frequency outside this stable area, selection actively drives one or more alleles to extinction. This phenomenon means it is more likely for genetic drift to cause allelic extinction with type II fitness sets rather than type I, since the random frequency change required to lead to extinction is smaller. This result shows the importance of genetic drift acting on well-established alleles after they have invaded the population; if genetic drift mainly prevented alleles from invading, the difference in relative proportion of type I and II fitness sets would not be so clear between populations with and without genetic drift. An alternative explanation is that type II fitness sets are harder to construct in smaller populations with genetic drift. This explanation seems unlikely to us, because genetic drift may actually aid invading alleles in reaching the particular allele frequencies that are part of the domain of attraction of locally stable equilibrium.
Table 1. The proportion of simulations leading either to type I, II or III fitness sets for three population sizes (N), seven levels of gene flow (m), with or without genetic drift.
| m | Type | 103 | 104 | 105 | |||
|---|---|---|---|---|---|---|---|
| Drift | No drift | Drift | No drift | Drift | No drift | ||
| 0 | I | 0.13 | 0.02 | 0.04 | 0.01 | 0.00 | 0.00 |
| II | 0.80 | 0.85 | 0.80 | 0.75 | 0.31 | 0.53 | |
| III | 0.07 | 0.13 | 0.16 | 0.24 | 0.69 | 0.47 | |
| 0.01 | I | 0.48 | 0.14 | 0.19 | 0.07 | 0.03 | 0.02 |
| II | 0.50 | 0.81 | 0.68 | 0.72 | 0.29 | 0.46 | |
| III | 0.02 | 0.05 | 0.13 | 0.21 | 0.68 | 0.52 | |
| 0.05 | I | 0.60 | 0.31 | 0.36 | 0.22 | 0.11 | 0.18 |
| II | 0.39 | 0.65 | 0.54 | 0.59 | 0.22 | 0.44 | |
| III | 0.02 | 0.05 | 0.10 | 0.19 | 0.67 | 0.38 | |
| 0.1 | I | 0.74 | 0.49 | 0.56 | 0.40 | 0.16 | 0.31 |
| II | 0.24 | 0.47 | 0.32 | 0.45 | 0.14 | 0.30 | |
| III | 0.02 | 0.04 | 0.12 | 0.16 | 0.68 | 0.39 | |
| 0.2 | I | 0.85 | 0.72 | 0.73 | 0.68 | 0.25 | 0.63 |
| II | 0.13 | 0.24 | 0.14 | 0.19 | 0.04 | 0.09 | |
| III | 0.02 | 0.05 | 0.13 | 0.14 | 0.71 | 0.28 | |
| 0.5 | I | 0.97 | 0.93 | 0.86 | 0.90 | 0.31 | 0.73 |
| II | 0.02 | 0.03 | 0.01 | 0.01 | 0.00 | 0.00 | |
| III | 0.01 | 0.04 | 0.13 | 0.10 | 0.69 | 0.27 | |
| 1.0 | I | 0.99 | 0.97 | 0.87 | 0.91 | 0.32 | 0.86 |
| II | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| III | 0.01 | 0.03 | 0.13 | 0.09 | 0.68 | 0.14 | |
| S | I | 0.99 | 0.95 | 0.87 | 0.84 | 0.32 | 0.68 |
| II | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| III | 0.01 | 0.05 | 0.13 | 0.16 | 0.68 | 0.32 |
Standard errors (calculated by bootstrapping 1000 replicates with a sample size of 1000) are all smaller than 0.016, with an average of 0.011. The letter S indicates the results for a single-deme model.
In contrast, for the largest population size (N = 105), genetic drift, rather than changing the relative proportion of type I/II fitness sets, substantially increases the proportion of type III fitness sets. This result is in line with the previous result from selection strength (s) that showed that in these large populations, genetic drift allows relatively more deleterious alleles to invade the population. Deleterious alleles will not be part of any selective equilibrium and therefore cause an increase in the proportion of type III fitness sets. Nevertheless, whereas the previous result concerning s suggested that deleterious alleles invaded mainly for high levels of m, when average fitness is lower, the effect of m on the proportion of type III fitness sets is limited. The stability analysis, however, detects only whether the recorded fully polymorphic set of alleles is maintained; this analysis does not detect the presence of multiple transient alleles. The combined result from both the stability and the strength of selection analysis, therefore, suggests the presence of multiple slightly deleterious alleles in the models with the highest N for highest levels of m. Overall, genetic drift increases the stability of polymorphic equilibria for N =103 and 104 compared to their non-drift equivalent models for most levels of gene flow, whereas the presence of transient alleles reduces the stability of these equilibria for N = 105.
Neutrality
We used the Ewens–Watterson test to investigate whether genetic drift and finite populations influence our ability to detect if the allele-frequency vectors resulting from our simulations differ from those generated by the neutral hypothesis. This test compares the observed level of homozygosity to that which is expected under neutrality. As a “backwards experiment” (Ramshaw et al. 1979), we sampled both allele-frequency vectors taken from a single deme and pooled frequency vectors taken from both demes to investigate the effect of correctly or incorrectly sampling the spatial structure.
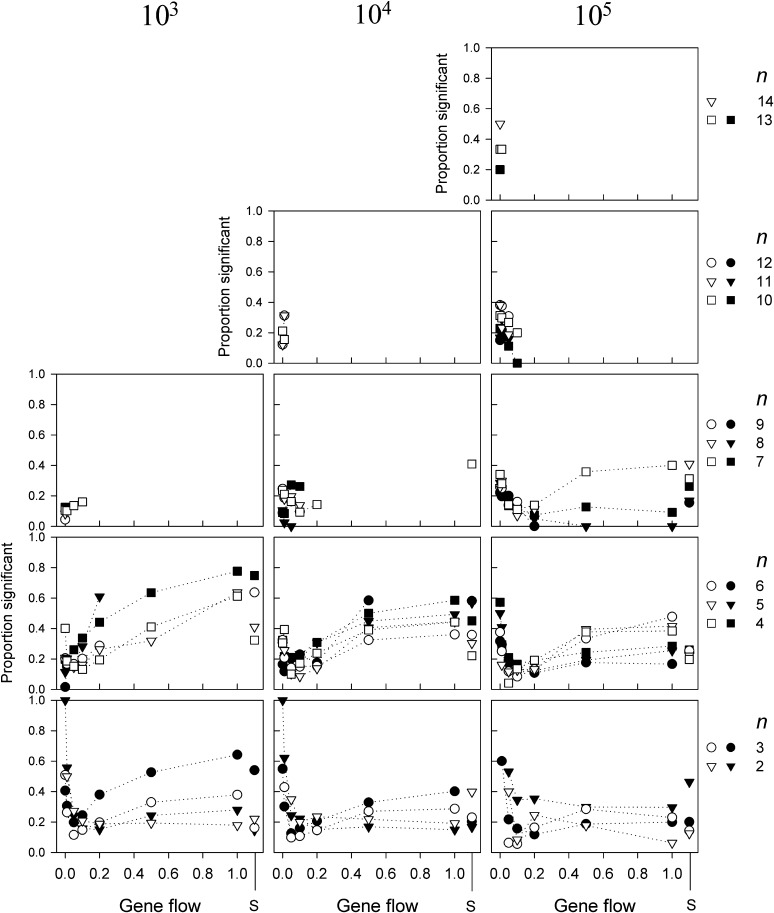
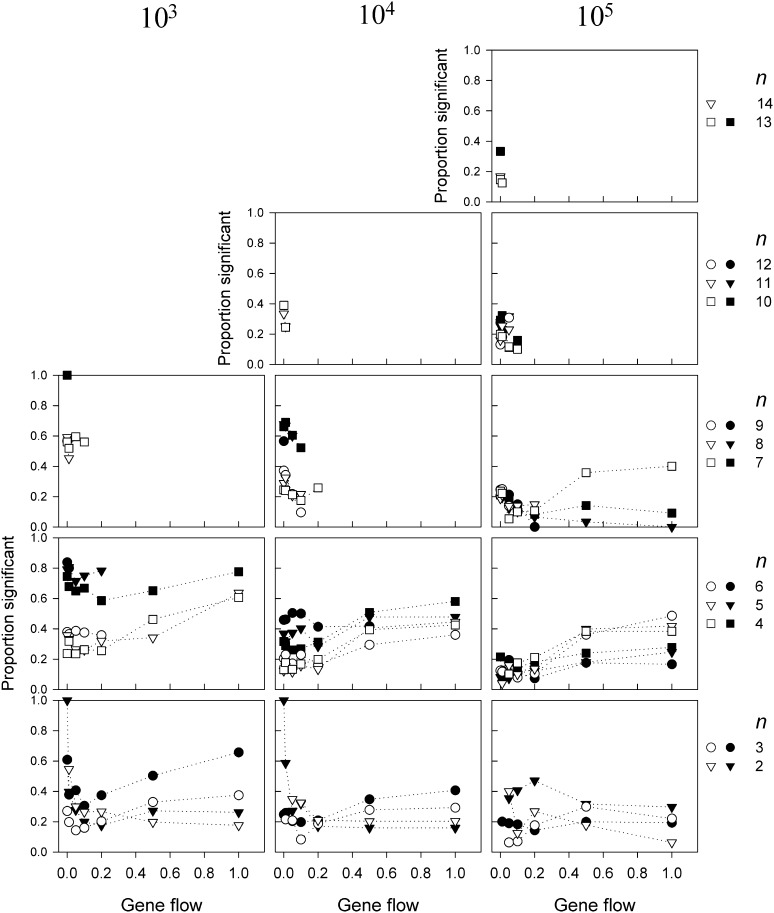
The ability to detect if allele-frequency vectors are significantly different from those expected under neutrality is rather low and, as in our previous investigations (e.g., Marks and Spencer 1991; Spencer and Marks 1992), for most simulation parameters the majority of vectors is not rejected. This ability to detect selection is further influenced by population sizes (N), gene flow (m), number of alleles (n), genetic drift, and whether the model was correctly sampled (Figure 4 and Figure 5). Some patterns do emerge, but these are not always consistent across all N, n, or in presence or absence of drift. For example, the proportion of allele-frequency vectors taken from a single deme that are detectably different from neutral is mostly lower for intermediate levels of gene flow but not always (e.g., not for N = 105, n = 8 with drift, and not for N = 103, n = 7, without drift). Similarly, most frequency vectors are found to be different from neutral for higher m in smaller populations, whereas more vectors for lower m in large populations, but not for n = 2. The fact that all frequency vectors for n = 2 and m = 0 are rejected as nonneutral, however, is an artifact; these vectors contain one allele at frequency 1.0 and another at frequency 0.0 (since each allele is present in only one of the demes). The observed level of homozygosity calculated from these samples is exactly 1.0, which is higher than the upper critical value for n = 2. Yet having two populations, each with a single allele, can hardly be considered proof of selection generating frequency patterns that are different from those generated by the neutral hypothesis. Finally, for higher m, genetic drift mostly increases the proportion of vectors detectably different from neutral for N = 103, 104, but mostly lowers this proportion for N = 105.
Figure 4.
The proportion of allele-frequency vectors sampled from one of the demes that were significantly different from neutral according to the Ewens–Watterson test for neutrality as a function of gene flow for three different population sizes N and n > 1. The solid symbols indicate the results for populations with random drift whereas the open symbols indicate those without. Allele-frequency vectors were rejected if 18 or more of the generated sample homozygosity values were outside the critical range of Ewens sampling distribution (see text for explanation). The letter S on the x-axis indicates the results for single-deme populations. Only combinations of N, m, and n are shown for which least 10 replicates were found.
Figure 5.
The proportion of pooled allele-frequency vectors that were significantly different from neutral according to the Ewens–Watterson test for neutrality as a function of gene flow for three different population sizes N and n > 1. The solid symbols indicate the results for populations with random drift whereas the open symbols indicate those without. Allele-frequency vectors were rejected if 18 or more of the generated sample homozygosity values were outside the critical range of Ewens sampling distribution (see text for explanation). Only combinations of N, m and n are shown for which at least 10 replicates were found.
For samples taken from pooled frequency vectors, genetic drift mostly increases the proportion of frequency vectors that are detectably different from neutral, especially for lower N and lower levels of gene flow (Figure 5). At these smaller population sizes, previous results show fewer rare alleles are present, which likely results in more equal frequency vectors and lower values of ; therefore, a larger proportion of frequency vectors is being rejected by the Ewens–Watterson test for being too even. In contrast, for high N and high m, settings for which previous results have shown a greater proportion of rare alleles, the proportion of frequency vectors being rejected is diminished in the population with genetic drift. This result suggests that these rare alleles increase the values of sufficiently to prevent detection due to equal distribution, but do not sufficiently increase the values of to allow detection due to skewed frequency distributions.
Similar to the results from allele-frequency vectors taken from a single deme, all pooled allele-frequency vectors for n = 2 and m = 0 are rejected as nonneutral. Again, this result is an artifact for having two different alleles in both demes. Averaging the allele frequencies for these settings results in a completely even frequency distribution, and calculated from these samples will be exactly 0.5, which is lower than the lower critical value for n = 2. Whereas for higher n and higher m, averaging of the allele-frequency vectors does not lead to completely even frequencies, frequency distributions will nonetheless be more equal compared to those from frequency vectors taken from a single deme. Thus, this averaging, ignoring spatial structure, causes more frequency vectors (which have already a more equal distribution for lower N due to fewer rare alleles) to be detected significantly different from neutral due to equal distribution of frequencies. While ignoring spatial structure appears to increase the ability to detect selection, this increased ability merely indicates that averaging allele-frequency distributions creates more even frequency distributions.
Discussion
The finiteness of populations has a considerable effect on the outcome on the construction of genetic polymorphism in our two-deme model. Two different aspects of finite populations can be recognized. First, genetic drift randomly changes allele frequencies from generation to generation (Wright 1937). This aspect has its most profound effects at low levels of gene flow, when effective population size in that deme is most reduced. The random frequency change can lead to the extinction of successful mutants and lowers the amount of genetic variation found in these smaller populations with genetic drift compared to those without genetic drift. This lower amount of genetic variation, however, in the presence of relatively strong levels of genetic drift is maintained in polymorphic equilibria that are on average more stable than those equilibria in similar sized populations without drift. This initially surprising result is easily explained: more stable polymorphic equilibria emerge from the models with genetic drift because this process can more easily move allele frequencies out of the smaller domains of attraction of less stable equilibria, driving those alleles out of the population. In other words, in a spatial model, sufficiently strong levels of genetic drift disproportionally affect the alleles that are not part of globally stable equilibria (or locally stable equilibria with large domains of attraction), leaving a subset of alleles that are. The emergence of more stable fitness sets is therefore a consequence of genetic drift, comparable to the emergence of heterosis as a consequence of natural selection (Ginzburg 1979).
Second, the finiteness of populations also has a major effect on absolute numbers of mutations that are introduced during the 104 generations and the allelic frequency at which they go extinct. The lower frequency at which alleles go extinct in larger populations increases their persistence time because these alleles are permitted to stay in the population longer (Marks and Spencer 1991). These longer persistence times allow more transient alleles to be present as population sizes increase. Both the higher numbers of mutations and the longer persistence time influence the dynamics of allele introduction and extinction.
Whereas genetic drift mostly lowers the amount of genetic variation found in our two deme model, counterintuitively the process increases variation for high levels of gene flow in the largest populations. This increase in amount of variation can be attributed to the invasion of slightly deleterious alleles. This result is surprising because larger (sub)populations are expected to have a lower level of deleterious alleles due to the effects of selection overwhelming the effects of genetic drift (Whitlock 2000; Theodorou and Couvet 2006). Genetic drift can introduce alleles that are slightly deleterious to populations either when drift is strong or when there are many mutations whose effect are small (Schultz and Lynch 1997; Whitlock 2003). Since genetic drift should be relatively less important for the largest population, our results suggest that the number of these mutations can be more important than the extent of the stochastic changes wrought by genetic drift. Moreover, we also find that the number of deleterious invasions increases with increasing levels of gene flow. For these higher levels of gene flow, the demes in our spatial model experience substantial levels of migration load, which decreases the average level of fitness. The lower average fitness may decrease the effectiveness by which slightly deleterious alleles are selected against, which may increase their persistence time.
Another, less obvious phenomenon may explain the higher number of slightly deleterious alleles. Whether a novel mutation is beneficial or deleterious depends on the average fitness in a particular population when that mutation arises (Whitlock 2000; Martin and Lenormand 2006). Indeed, a critical feature of our construction model is that the rate of beneficial to deleterious alleles is not constant over the 104 generations. As alleles accumulate, average levels of fitness increase, which increases the proportion of deleterious mutations. After many generations, most mutations will be deleterious, some slightly deleterious, and almost none beneficial. How many mutations can be considered to fall in the range of slightly deleterious, therefore, does not depend only on the distribution of mutational fitnesses (Schultz and Lynch 1997; Whitlock 2003; Rodriguez-Ramilo et al. 2004), but also on the average level of fitness that is achieved after many generations of mutation and selection. Consequently, these construction models seem to evolve to be somewhat comparable to the nearly neutral model (Ohta 1992) and are exposed to a considerable number of slightly deleterious alleles, especially for the larger population sizes that encounter a higher number of new mutants. Perhaps the lower level of average fitness due to migration load further increases the number of mutations that are slightly deleterious. Such increased exposure to slightly deleterious alleles could increase the probability of their invasion.
The allele-frequency patterns resulting from these models are influenced by genetic drift, migration, sampling from a single deme or pooled, and finally selection. Because selection is only one of these factors, and the frequency patterns are easily confounded by these other factors, it is not surprising that the Ewens–Watterson test (Ewens 1972; Watterson 1977, 1978), designed to detect selection based on allele-frequency data, has a low power, especially in a spatial model (Gillespie 1991; Star et al. 2007a). Nevertheless, genetic drift and finite populations have some interesting effects on the proportion of frequency vectors that can be distinguished from neutrality. For smaller populations, the combined effect of genetic drift and a lower number of mutants encountered results in a reduction of rare alleles. Therefore, the remaining alleles are likely to have a more even frequency distribution, which results in more vectors being rejected from neutrality for being considered too even. This effect is especially strong when spatial structure is ignored, and all frequencies are pooled. In contrast, for larger populations, the higher number of mutants encountered results in more rare alleles and more skewed frequency distributions. Genetic drift further increases this number of rare alleles by the introduction of slightly deleterious alleles. While the increase in skewed frequency distributions obviously reduces the number of allele-frequency vectors that can be distinguished for being too even, it does not sufficiently increase the skew of the frequency vectors enough to increase the number of rejections for being too skewed. Interestingly, a random process like genetic drift can appear to improve the power of a test to detect selection by eliminating rare alleles and decrease this power by introducing these rare alleles for different levels of gene flow. This result highlights the limitations of the traditional interpretation that rejection from neutrality due to even distributions is evidence for selection for heterozygotes and rejection due to skewed distribution evidence for selection against heterozygotes (Manly 1985; Marks and Spencer 1991).
Overall, genetic drift has two profoundly different effects on the construction of genetic polymorphisms in our spatial model. Interestingly, both effects do not occur simultaneously; genetic drift drives relatively more alleles toward extinction for smaller populations as expected, whereas it introduces slightly deleterious alleles for larger populations. These different effects can effectively decrease or increase the levels of genetic variation found at the end of our simulations and further influence the level of adaptation, the stability of the polymorphisms, and whether the allele-frequency vectors can be distinguished from those expected under selective neutrality.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their comments and suggestions. This manuscript benefited greatly from discussions with Meredith V. Trotter. The genetic drift algorithm was kindly provided by Rick J. Stoffels. This work was supported by the Allan Wilson Centre for Molecular Ecology and Evolution and the University of Otago Postgraduate Scholarships.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Bonneuil N., 2012. Multiallelic polymorphism maintained under unpredictable migration and selection. J. Theor. Biol. 293: 189–196. [DOI] [PubMed] [Google Scholar]
- Bürger R., 2009. Polymorphism in the two-locus Levene model with nonepistatic directional selection. Theor. Popul. Biol. 76: 214–228. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1972. Sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1976. The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 10: 253–280. [DOI] [PubMed] [Google Scholar]
- Gentle J. E., 2003. Random Number Generation and Monte Carlo Methods. Springer-Verlag, New York. [Google Scholar]
- Gillespie J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, New York. [Google Scholar]
- Ginzburg L. R., 1979. Why are heterozygotes often superior in fitness? Theor. Popul. Biol. 15: 264–267. [Google Scholar]
- Hall M. C., Willis J. H., 2006. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60: 2466–2477. [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 2007. Principles of Population Genetics, Ed. 4 Sinauer, Sunderland. [Google Scholar]
- Hedrick P. W., Ginevan M. E., Ewing E. P., 1976. Genetic-polymorphism in heterogeneous environments. Annu. Rev. Ecol. Syst. 7: 1–32. [Google Scholar]
- Holst L., 1980. On the lengths of the pieces of a stick broken at random. J. Appl. Probab. 17: 623–634. [Google Scholar]
- Labbe P., Lenormand T., Raymond M., 2005. On the worldwide spread of an insecticide resistance gene: a role for local selection. J. Evol. Biol. 18: 1471–1484. [DOI] [PubMed] [Google Scholar]
- Leimu R., Fischer M., 2008. A meta-analysis of local adaptation in plants. PLoS ONE 3: e4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., 1974. The Genetic Basis of Evolutionary Change, Columbia University Press, New York. [Google Scholar]
- Lewontin R. C., Ginzburg L. R., Tuljapurkar S. D., 1978. Heterosis as an explanation for large amounts of genic polymorphism. Genetics 88: 149–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B. F. J., 1985. The Statistics of Natural Selection, Chapman & Hall, London. [Google Scholar]
- Marks R. W., Spencer H. G., 1991. The maintenance of single-locus polymorphism. 2. The evolution of fitnesses and allele frequencies. Am. Nat. 138: 1354–1371. [Google Scholar]
- Martin G., Lenormand T., 2006. The fitness effect of mutations across environments: a survey in light of fitness landscape models. Evolution 60: 2413–2427. [PubMed] [Google Scholar]
- Ohta T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263–286. [Google Scholar]
- Ohta T., Gillespie J. H., 1996. Development of neutral and nearly neutral theories. Theor. Popul. Biol. 49: 128–142. [DOI] [PubMed] [Google Scholar]
- Peischl S., 2010. Dominance and the maintenance of polymorphism in multiallelic migration-selection models with two demes. Theor. Popul. Biol. 78: 12–25. [DOI] [PubMed] [Google Scholar]
- Ramshaw J. A. M., Coyne J. A., Lewontin R. C., 1979. The sensitivity of gel-electrophoresis as a detector of genetic-variation. Genetics 93: 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ramilo S. T., Perez-Figueroa A., Fernandez B., Fernandez J., Caballero A., 2004. Mutation-selection balance accounting for genetic variation for viability in Drosophila melanogaster as deduced from an inbreeding and artificial selection experiment. J. Evol. Biol. 17: 528–541. [DOI] [PubMed] [Google Scholar]
- Schultz S. T., Lynch M., 1997. Mutation and extinction: the role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51: 1363–1371. [DOI] [PubMed] [Google Scholar]
- Spencer H. G., Marks R. W., 1988. The maintenance of single-locus polymorphism. 1. Numerical-studies of a viability selection model. Genetics 120: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer H. G., Marks R. W., 1992. The maintenance of single-locus polymorphism. 4. Models with mutation from existing alleles. Genetics 130: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer H. G., Marks R. W., 1993. The evolutionary construction of molecular polymorphisms. N. Z. J. Bot. 31: 249–256. [Google Scholar]
- Star B., Stoffels R. J., Spencer H. G., 2007a Evolution of fitnesses and allele frequencies in a population with spatially heterogeneous selection pressures. Genetics 177: 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B., Stoffels R. J., Spencer H. G., 2007b Single-locus polymorphism in a heterogeneous two-deme model. Genetics 176: 1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B., Trotter M. V., Spencer H. G., 2008. Evolution of fitnesses in structured populations with correlated environments. Genetics 179: 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffels R. J., Spencer H. G., 2008. An asymmetric model of heterozygote advantage at major histocompatibility complex genes: degenerate pathogen recognition and intersection advantage. Genetics 178: 1473–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P. J., 1985 Construction and turnover of multispecies communities: a critique of approaches to ecological complexity. Ph.D. Thesis, Harvard University, Cambridge. MA.
- Theodorou K., Couvet D., 2006. Genetic load in subdivided populations: interactions between the migration rate, the size and the number of subpopulations. Heredity 96: 69–78. [DOI] [PubMed] [Google Scholar]
- Trotter M. V., Spencer H. G., 2008. The generation and maintenance of genetic variation by frequency-dependent selection: constructing polymorphisms under the pairwise interaction model. Genetics 180: 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1977. Heterosis or neutrality. Genetics 85: 789–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1978. Homozygosity test of neutrality. Genetics 88: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., 2000. Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles, and sexual selection. Evolution 54: 1855–1861. [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2003. Fixation probability and time in subdivided populations. Genetics 164: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1937. The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.