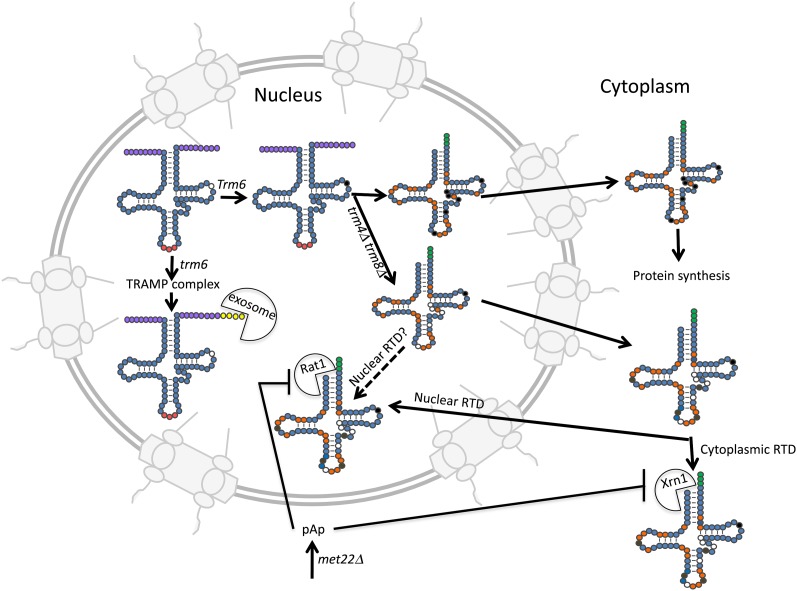
Figure 5.
tRNA turnover pathways in S. cerevisiae. Initial tRNA transcripts with 5′ and 3′ extensions (purple circles) are substrates for 3′ to 5′ exonucleolytic degradation by the nuclear exosome if the transcripts are missing a particular modification, m1A58 (open circle on initial tRNA transcript) or if 3′ processing is aberrant (not shown). The unmodified/aberrant tRNAs first receive A residues at the 3′ end (yellow circles) by the activity of the TRAMP complex and then the tRNAs are degraded by the nuclear exosome (exosome pac-man). Aberrant tRNAs can also be degraded by the rapid tRNA turnover pathway (RTD) in either the nucleus or the cytoplasm. The RTD pathway acts upon tRNAs missing particular multiple modifications or tRNAs that are otherwise unstructured (see text). As an example, tRNAs missing multiple modifications (open circles) due to trm4Δ trm8Δ mutations are subject to 5′ to 3′ degradation by the exonucleases in either the nucleus (RAT1 pac-man) or in the cytoplasm (Xrn1 pac-man). Solid circles are those modifications affected by mutations of TRM6 or TRM4 and TRM8. Orange circles indicate modifications acquired in the nucleus, whereas brown circles indicate modifications acquired in the cytoplasm. CCA nucleotides are indicated by green circles and the anticodon by red circles. Also indicated is pAp, an intermediate of methionine biosynthesis that inhibits Xrn1 and Rat1, thereby connecting tRNA turnover to amino acid biosynthesis.