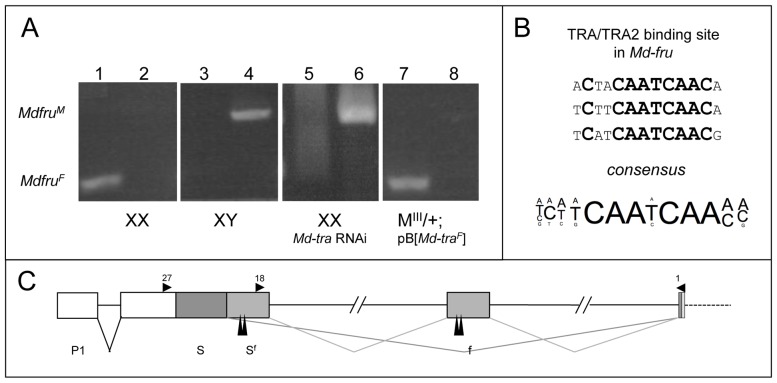
Figure 2. Sex-specific splicing of Md-fru is mediated by Md-tra.
(A) RT-PCR results with sex-specific sets of Md-fru primer pairs. Amplification with Md-fruF specific primers Md-fru-18 and Md-fru-1 in wildtype XX females (1), in wildtype XY males (3), in Md-tra silenced XX females (5) and in ectopically Md-traF expressing MIII males (7). Amplification with Md-fruM specific primers Md-fru-27 and Md-fru-1 in wildtype XX females (2), in wildtype XY males (4), in Md-tra silenced XX females (6) and in ectopically Md-traF expressing MIII males (8). In wildtype XX flies, where Md-tra is active, only Md-fruF but no Md-fruM transcripts can be detected (1 and 2). A clear shift to Md-fruM splicing can be observed in XX individuals when Md-tra is silenced (5 and 6). Likewise, only Md-fruM transcripts are present in wildtype XY males, where Md-tra is not active (3 and 4). Md-fru splicing shifts to the female mode in MIII males when continuous Md-traF activity is provided by a transgene [28] (7+8). (B) Direct splicing regulation by Md-tra is suggested by the presence of putative TRA/TRA2 binding sites in Md-fru. Comparison of three different sequences found in Md-fru with the TRA/TRA2 binding site consensus derived from sequence alignments of binding sites found in the fru and dsx orthologs of Drosophila melanogaster, Musca, domestica and Anopheles gambiae. (C) Putative TRA/TRA2 binding sites (long vertical arrowheads) are found in close vicinity to the regulated splicing sites in the Md-fru locus. Positions of the primers used in this expression analysis are indicated by small horizontal arrowheads.