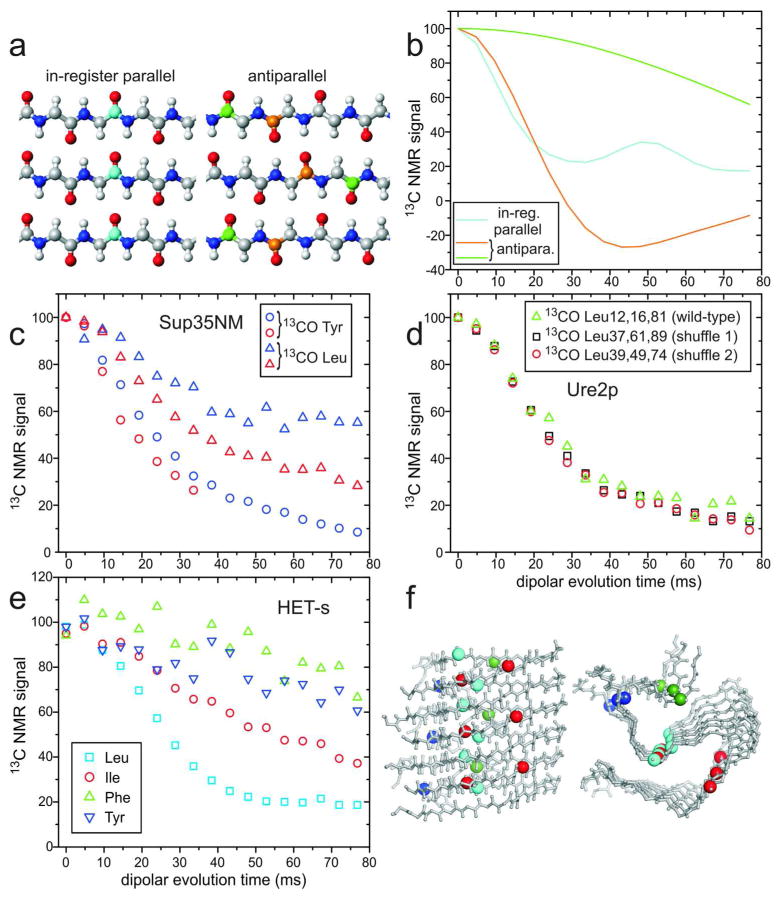
Figure 5.
Investigations of β-sheet structures in prion fibrils, through measurements of intermolecular 13C-13C magnetic dipole couplings with the PITHIRDS-CT technique.20 (a) Ideal in-register parallel and antiparallel β-sheets. (b) Ideal PITHIRDS-CT curves for an in-register parallel β-sheet with a single carbonyl 13C label at any residue in each peptide chain (cyan) and for antiparallel β-sheets with a single carbonyl 13C label at either of two residues (orange and green). (c) PITHIRDS-CT data for Sup35NM fibrils, prepared with 13C labels at backbone carbonyl sites of all Tyr (circles) or Leu (triangles) residues. Data were obtained from lyophilized (blue) or fully hydrated (red) samples.41 All 20 Tyr residues are in the N domain of Sup35NM, while 7 of 8 Leu residues are in the M domain. (d) PITHIRDS-CT data for wild-type Ure2p1–89 fibrils (triangles), prepared with 13C labels at backbone carbonyl sites of all Leu residues, and for two “shuffled prion” fibrils40 with Leu at the indicated residue numbers (squares and circles). (e) PITHIRDS-CT data for HET-s fibrils, residues 218–289, prepared with 13C labels at backbone carbonyl sites of the indicated residues, in separate samples. (f) Side and cross-sectional views of the HET-s fibril model developed by van Melckebeke et al. (PDB 2KJ3).4 Residues 224–248 and 259–289 are shown, corresponding to the most highly ordered and rigid segments. Labeled carbonyl sites are represented by large spheres, colored according to panel e. In the β-helical core structure of HET-s fibrils, only Leu carbonyls have ~5 Å nearest-neighbor distances. Signal contributions from labeled sites outside the core are uncertain. (All PITHIRDS-CT data are corrected for natural-abundance 13C contributions.41)