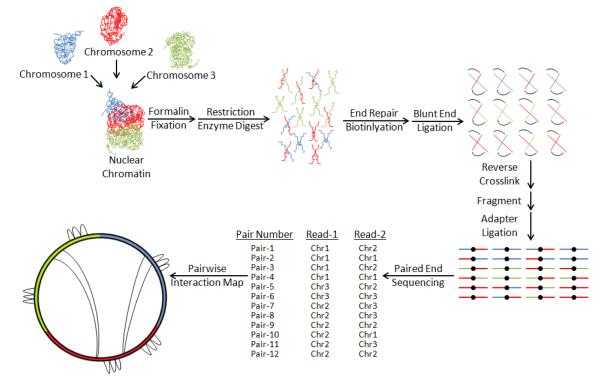
Figure 2. HiC Method for Chromosomal Conformation Sequencing.
Long-range interchromosomal and intrachromosomal interactions implicated in regulation of epigenetically coordinated gene networks can be identified by HiC. Briefly, chromosomal interactions are preserved by formalin fixation, while restriction enzyme digestion cuts the genome into fragments containing sites of formalin fixation between genomic segments. Sticky ends are then repaired using biotinylated nucleotides and the repaired ends are ligated as blunt-ends introducing biotin at the site of ligation before crosslinks are reversed. DNA is then sheared to a fragment size compatible with high-throughput sequencing, biotin bearing fragments are isolated, and a paired-end sequencing library is made where each end represents a partner in a pairwise interaction. Pairs are then mapped by sequence alignment to a reference genome, creating a genome-wide atlas of structural interactions.