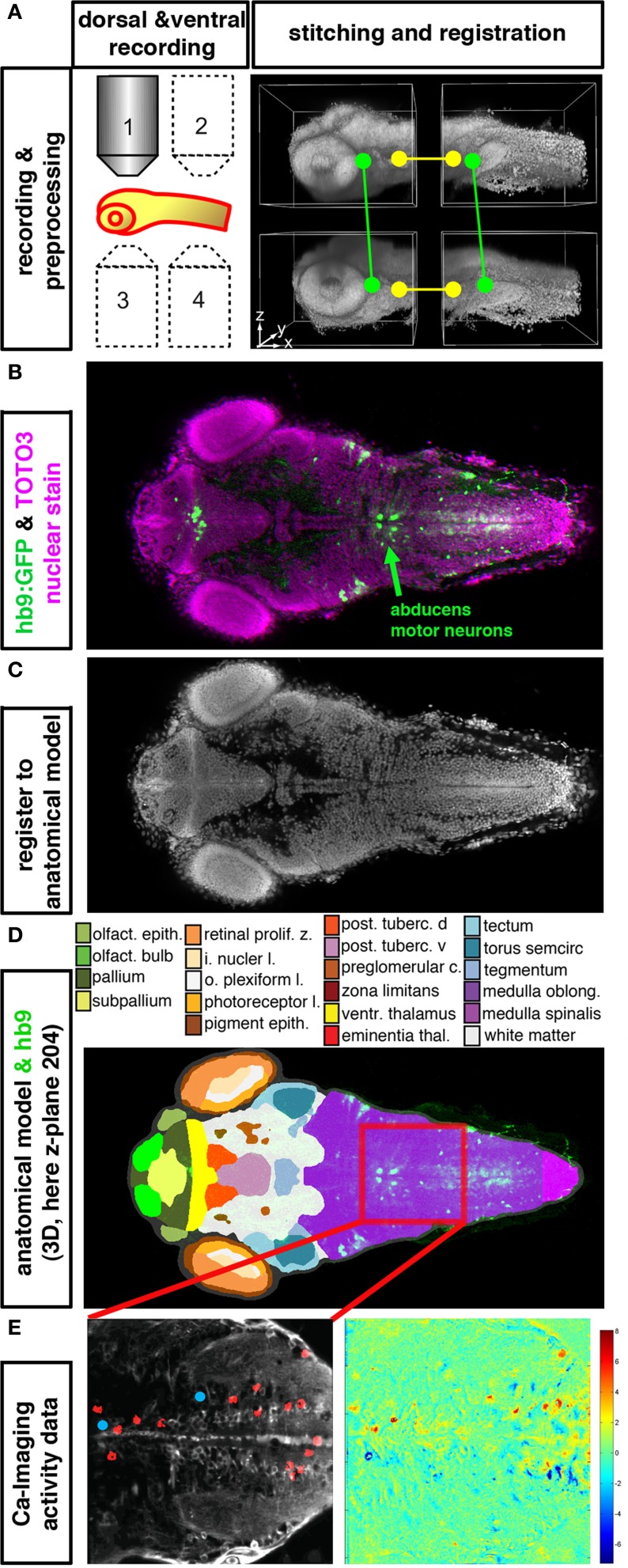
Figure 2.
Integration of anatomical, gene expression, and functional information in 3D. (A–E) High resolution datasets generated using an anatomical reference stain for each developmental stage may serve to establish an anatomical reference to register different data types into an anatomical and functional atlas of the nervous system. The Virtual Brain Explorer for Zebrafish (ViBE-Z) provides a framework for generation of high resolution 3D image stacks and for registration into an anatomical model (Ronneberger et al., 2012). (A) For single cell resolution documentation of the whole zebrafish brain, image stacks of rostral and caudal parts of the brain are recorded from ventral and dorsal sides using standard single photon confocal microscopes. Fluorescent stain of all cell nuclei is used to obtain anatomical information, and also to correct for loss of signal in deep tissues, assuming that all nuclei contain the same amount of DNA and have similar stain intensities. ViBE-Z stitches the individual stacks into one high resolution 3D data volume and performs correction of optical attenuation (light absorption and scattering in tissue). (B) Example of a ViBE-Z processed dataset of a 3-day-old larva showing GFP expression from Tg(hb9:GFP) (Flanagan-Steet et al., 2005) and TOTO3 fluorescent nuclear stain. (C) The fluorescent stain of cell nuclei is used by ViBE-Z to extract landmark information and register the experimental datasets in 3D to a reference larva. (D) The anatomical model of the reference embryo may now be combined with the experimental data set. Here, a dorsal view at focal plane z_204 of the reference larva is shown. The experimental Tg(hb9:GFP) expression provides neuronal information even in the medulla oblongata hindbrain region, which has only few anatomical annotations in reference databases (www.ZFIN.org). (E) In the future, as soon as anatomical references may be recorded also in live larva, the anatomical and gene expression information in ViBE-Z may be used to identify neurons which have defined recorded activity patterns. In this case, neurons with activity patterns correlating with oculomotor activity have been detected (see Figure 1), and ViBE-Z may be used to determine whether some of these active neurons may correspond to abducens motor neurons, which are labeled in the Tg(hb9:GFP) transgenic line.