Fig 1.
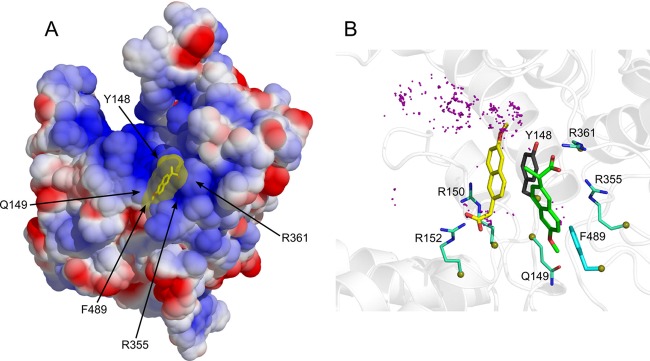
(A) Binding of naproxen to the RNA binding groove of the nucleoprotein from influenza A H1N1 virus based on PDB 2IQH (5). The protein surface is shown according to the electrostatic potential (blue, positive potential arising from the multiple arginine and lysine residues in the RNA binding groove; red, negative potential). This structure of the NP-naproxen complex solvated by water molecules was obtained after 10 ns of MD simulations. Details of the interactions of naproxen with NP inserted in a small hydrophobic cavity defined by Y148 and F489 are shown in panel B. (B) Superposition of the NP-naproxen complexes with the lowest energy. Virtual screening was used to define the possible binding site(s) of naproxen in the RNA binding groove of NP. The most MD-stable structure of the initial naproxen structure (whose carbon atoms are colored in green) is stabilized by hydrophobic interactions with Y148 and F489, a salt bridge with R361, and a water-mediated salt bridge with R355. Y148 stacks on the naphthalene core of naproxen, and the methoxy group of naproxen often forms an H-bond interaction with Q149 (see also Fig. S1 and Movie S6 in the supplemental material). The naproxen was further docked on NP using a searching volume extended to the rigid RNA binding site. The docking poses are represented as purple dots. The naproxen structure (with carbon atoms colored in yellow) with the lowest interaction energy (electrostatic and van der Waals) made a salt bridge with R152, a water-mediated salt bridge with R150, and a π-π interaction with Y148.
