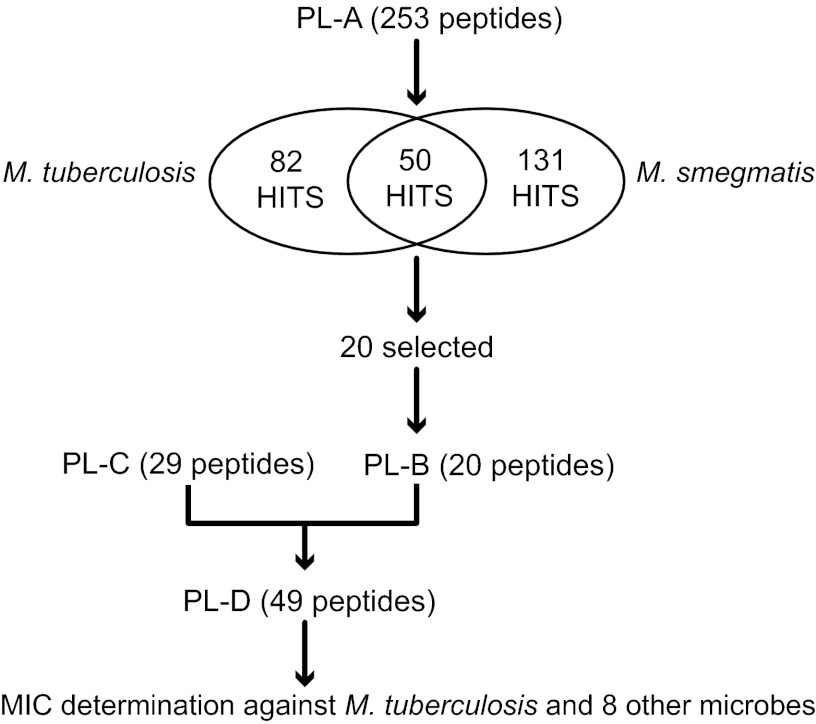
Fig 1.
Flowchart of the screening program used to select antimycobacterial peptides. Peptide library A (PL-A) was composed of 253 peptides from several published (19–22) and unpublished libraries, chosen for their antibacterial activities against Pseudomonas aeruginosa (note that 214 of these peptides qualified as positive hits against P. aeruginosa by the more stringent criteria used in previous studies [24]. The 39 nonactives were introduced as internal negative controls). Peptides in PL-A were screened against M. smegmatis and M. tuberculosis. Those active against M. tuberculosis, representing a range of amino acid compositions and sequences, were chosen for further analyses as PL-B (20 peptides). This pool was combined with 29 peptides selected for activities against P. aeruginosa and other representative pathogenic microbes (PL-C; CAMPs active against P. aeruginosa [unpublished data]) to generate PL-D. The antimicrobial activities of the 49 peptides in PL-D were quantified against a broad range of pathogenic microbes.