Abstract
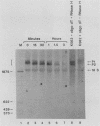
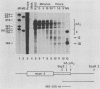
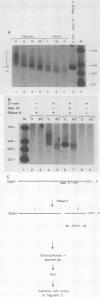
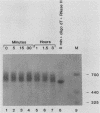
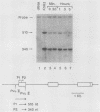
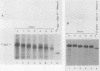
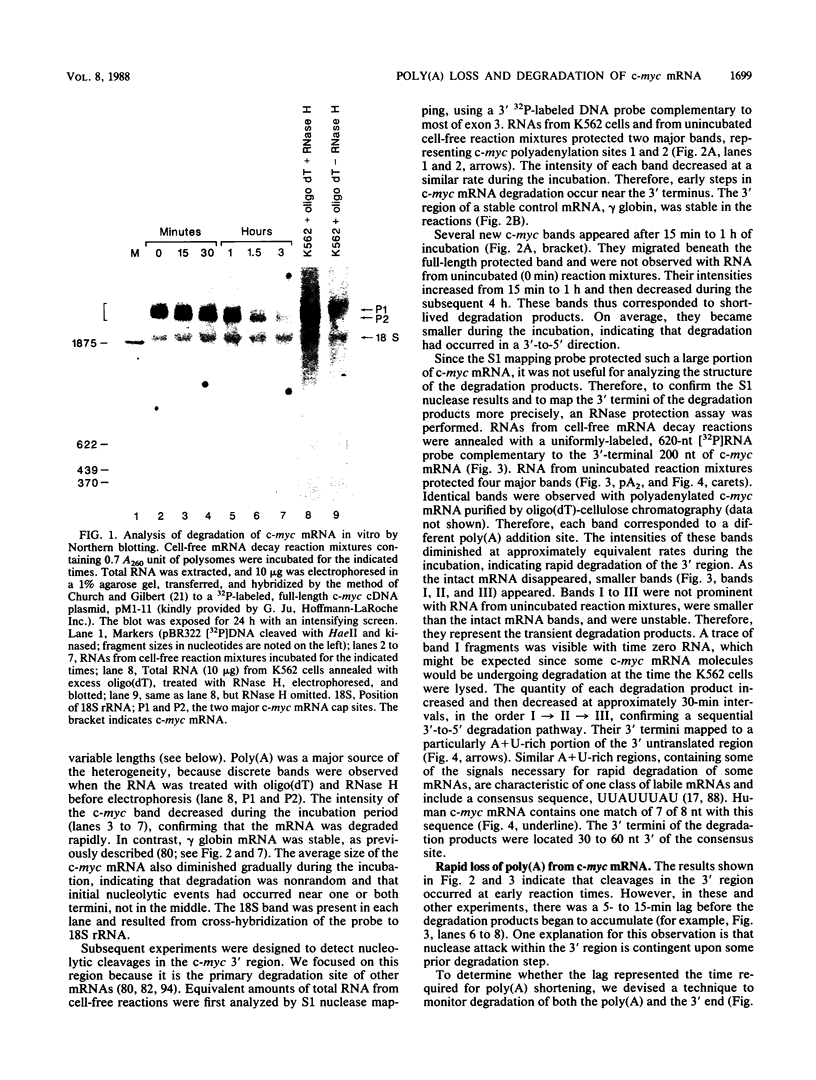
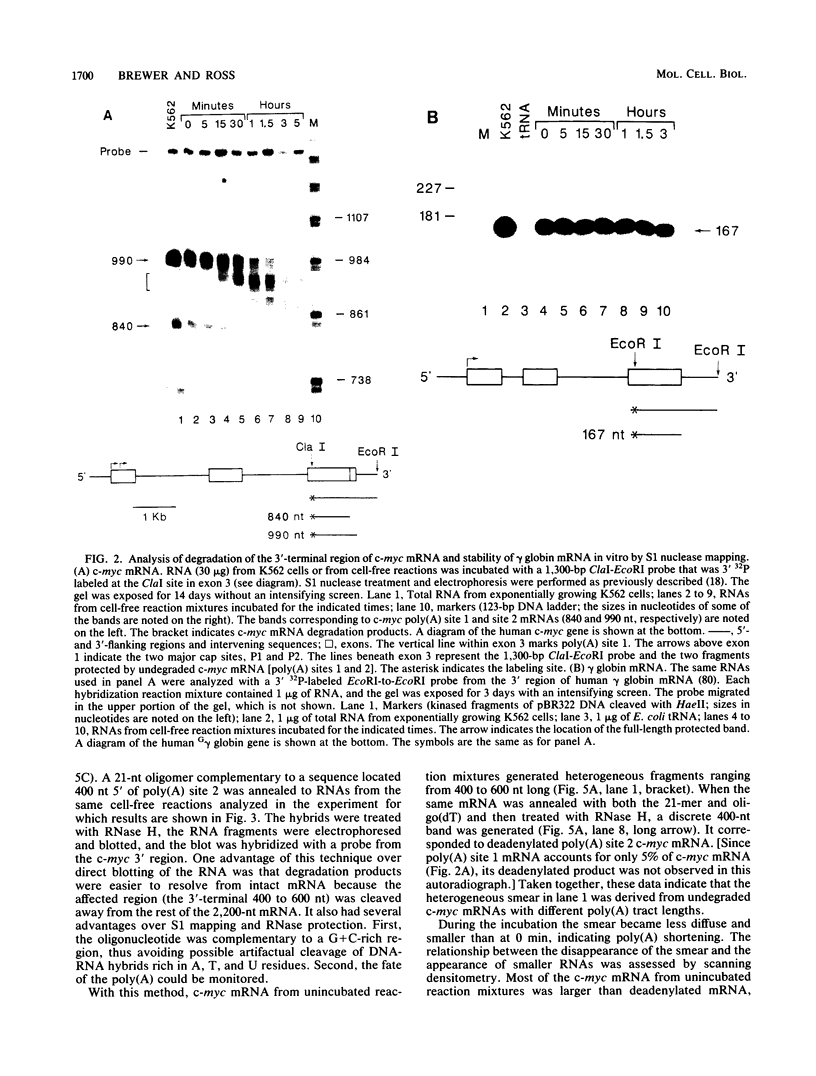
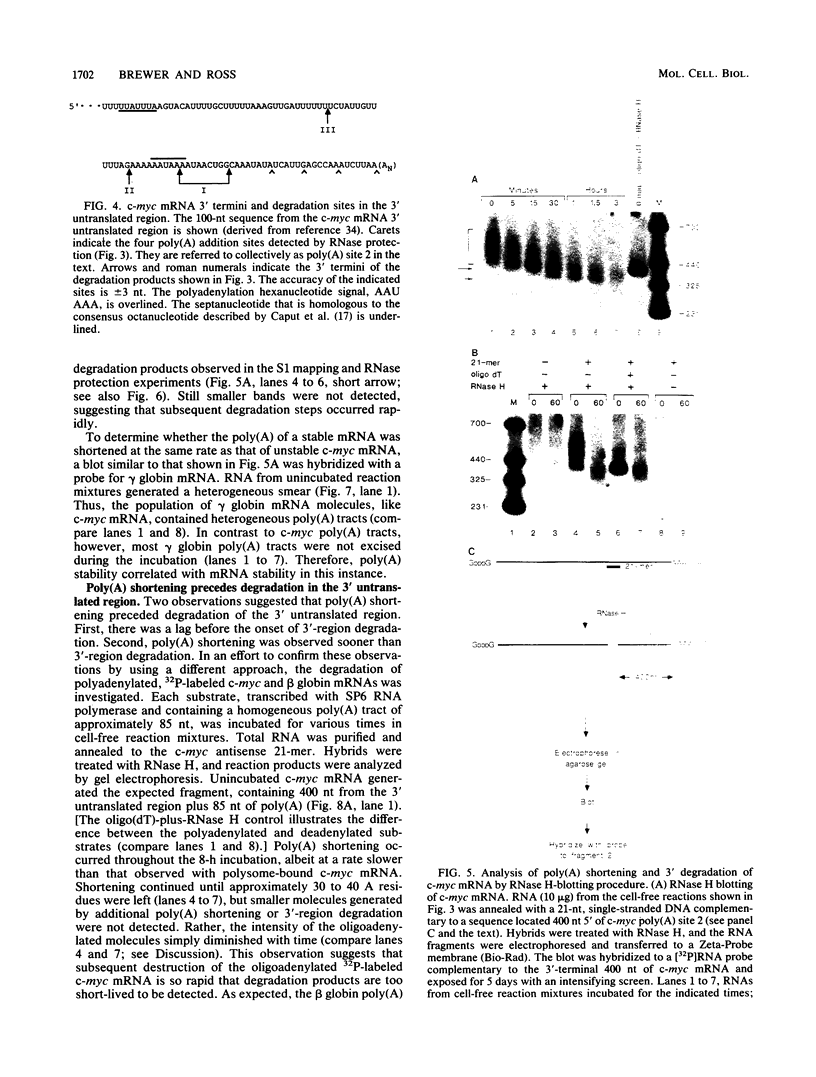
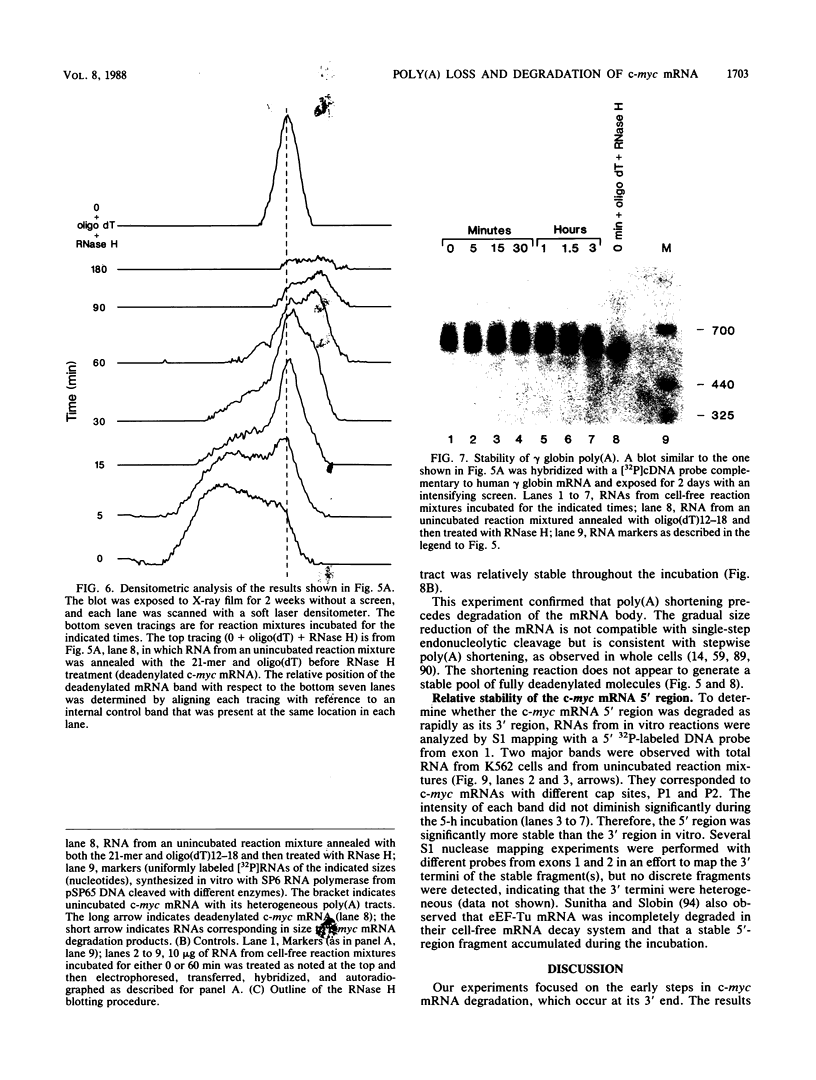
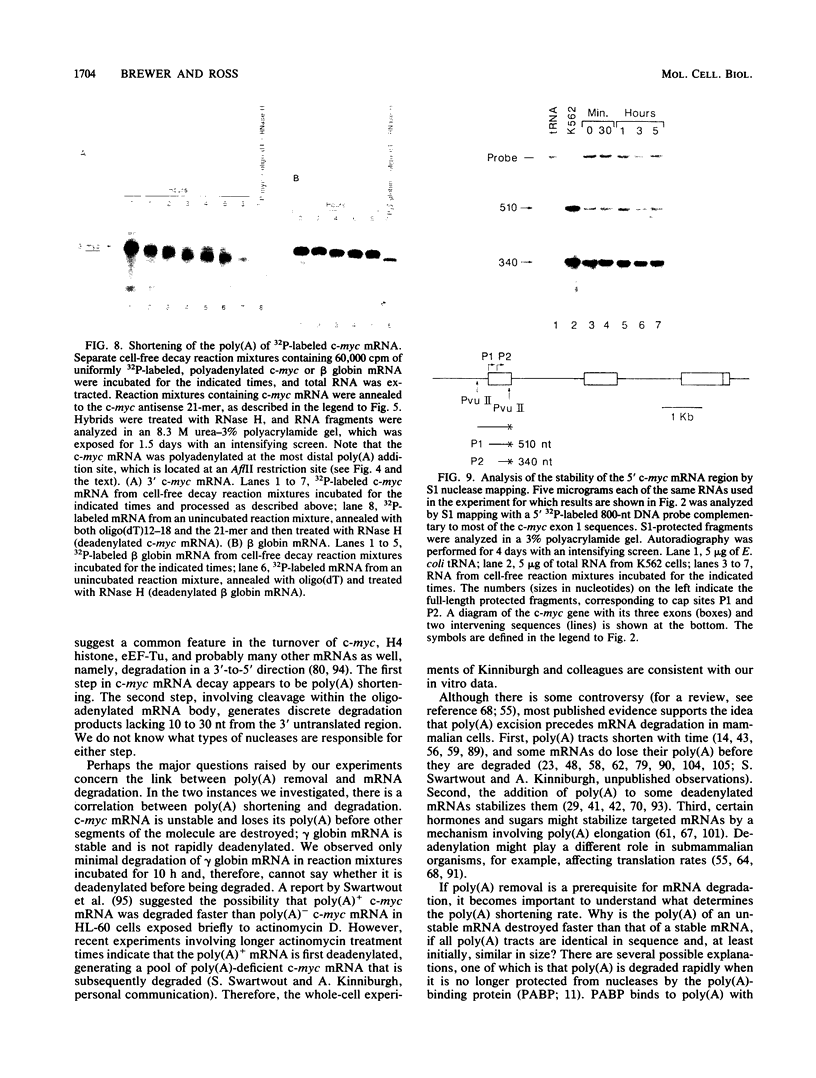
The early steps in the degradation of human c-myc mRNA were investigated, using a previously described cell-free mRNA decay system. The first detectable step was poly(A) shortening, which generated a pool of oligoadenylated mRNA molecules. In contrast, the poly(A) of a stable mRNA, gamma globin, was not excised, even after prolonged incubation. The second step, degradation of oligoadenylated c-myc mRNA, generated decay products whose 3' termini were located within the A+U-rich portion of the 3' untranslated region. These products disappeared soon after they were formed, consistent with rapid degradation of the 3' region. In contrast, the 5' region, corresponding approximately to c-myc exon 1, was stable in vitro. The data indicate a sequential degradation pathway in which 3' region cleavages occur only after most or all of the poly(A) is removed. To account for rapid deadenylation, we suggest that the c-myc poly(A)-poly(A)-binding protein complex is readily dissociated, generating a protein-depleted poly(A) tract that is no longer resistant to nucleases.
Full text
PDF

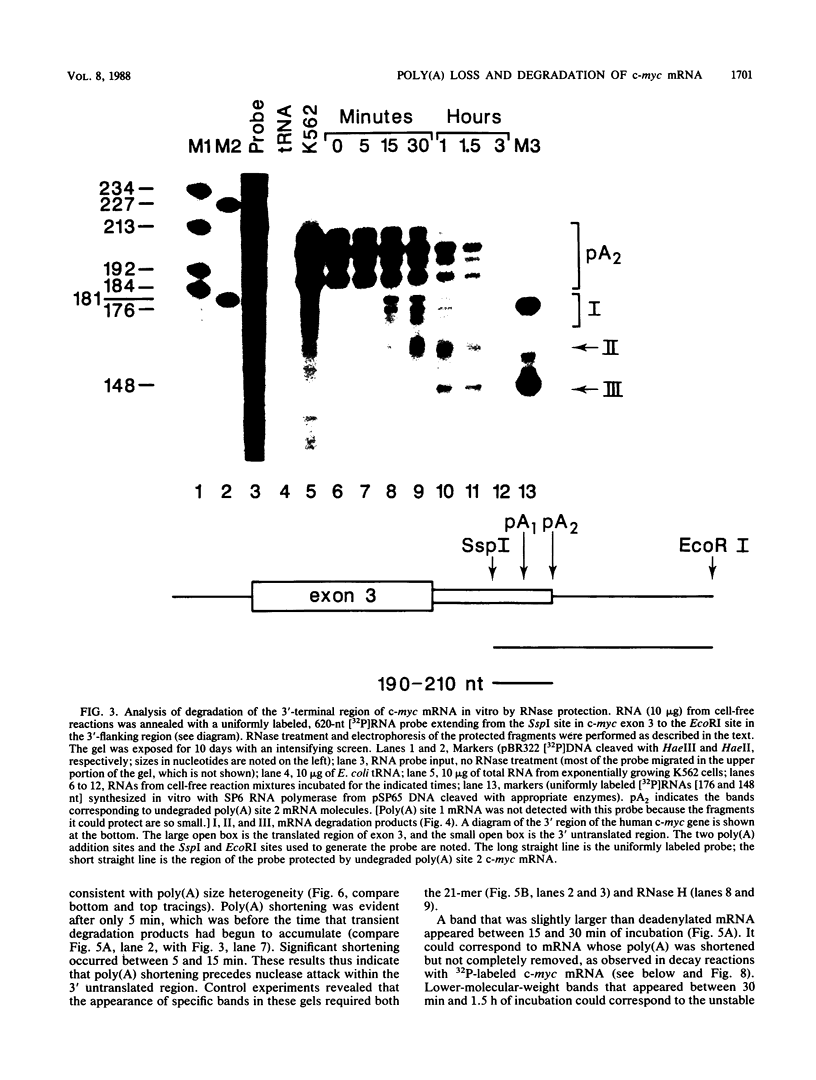




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alitalo K., Ramsay G., Bishop J. M., Pfeifer S. O., Colby W. W., Levinson A. D. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983 Nov 17;306(5940):274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Kornberg R. D. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Kornberg R. D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983 Mar;96(3):717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann I. E., Brawerman G. Control of breakdown of the polyadenylate sequence in mammalian polyribosomes: role of poly(adenylic acid)-protein interactions. Biochemistry. 1977 Jan 25;16(2):259–264. doi: 10.1021/bi00621a016. [DOI] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Blobel G. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc Natl Acad Sci U S A. 1973 Mar;70(3):924–928. doi: 10.1073/pnas.70.3.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G., Diez J. Metabolism of the polyadenylate sequence of nuclear RNA and messenger RNA in mammalian cells. Cell. 1975 Jul;5(3):271–280. doi: 10.1016/0092-8674(75)90102-6. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson D. P., Ross J. Human beta-globin promoter and coding sequences transcribed by RNA polymerase III. Cell. 1983 Oct;34(3):857–864. doi: 10.1016/0092-8674(83)90543-3. [DOI] [PubMed] [Google Scholar]
- Carneiro M., Schibler U. Accumulation of rare and moderately abundant mRNAs in mouse L-cells is mainly post-transcriptionally regulated. J Mol Biol. 1984 Oct 5;178(4):869–880. doi: 10.1016/0022-2836(84)90316-4. [DOI] [PubMed] [Google Scholar]
- Chodchoy N., Levine B. J., Sprecher C., Skoultchi A. I., Marzluff W. F. Expression of mouse histone genes: transcription into 3' intergenic DNA and cryptic processing sites downstream from the 3' end of the H3 gene. Mol Cell Biol. 1987 Mar;7(3):1039–1047. doi: 10.1128/mcb.7.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby W. W., Chen E. Y., Smith D. H., Levinson A. D. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature. 1983 Feb 24;301(5902):722–725. doi: 10.1038/301722a0. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Rosbash M. Behavior of individual maternal pA+ RNAs during embryogenesis of Xenopus laevis. Dev Biol. 1982 Nov;94(1):79–86. doi: 10.1016/0012-1606(82)90070-7. [DOI] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A., Ley T. J., Humphries R. K., Fordis M., Schechter A. N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Campisi J. c-myc regulation during retinoic acid-induced differentiation of F9 cells is posttranscriptional and associated with growth arrest. Mol Cell Biol. 1986 Feb;6(2):518–524. doi: 10.1128/mcb.6.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. K., Chatterjee B., Roy A. K. Translation and stability of rat liver messenger RNA for alpha 2 mu-globulin in Xenopus oocyte. The role of terminal poly(A). J Biol Chem. 1979 Sep 25;254(18):8937–8942. [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C., Rigolet M., Briand J. P., Van Regenmortel M. H., Galibert F. Immunochemical detection of proteins related to the human c-myc exon 1. EMBO J. 1986 Sep;5(9):2241–2250. doi: 10.1002/j.1460-2075.1986.tb04491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeau F., Persson H., Gray H. E., Pardee A. B. C-myc expression is dissociated from DNA synthesis and cell division in Xenopus oocyte and early embryonic development. EMBO J. 1986 Dec 20;5(13):3571–3577. doi: 10.1002/j.1460-2075.1986.tb04684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyette M., Petropoulos C. J., Shank P. R., Fausto N. Regulated transcription of c-Ki-ras and c-myc during compensatory growth of rat liver. Mol Cell Biol. 1984 Aug;4(8):1493–1498. doi: 10.1128/mcb.4.8.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppi V. E., Jr, Coffino P. G1 and S phase mammalian cells synthesize histones at equivalent rates. Cell. 1980 Aug;21(1):195–204. doi: 10.1016/0092-8674(80)90127-0. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huez G., Marbaix G., Gallwitz D., Weinberg E., Devos R., Hubert E., Cleuter Y. Functional stabilisation of HeLa cell histone messenger RNAs injected into Xenopus oocytes by 3'-OH polyadenylation. Nature. 1978 Feb 9;271(5645):572–573. doi: 10.1038/271572a0. [DOI] [PubMed] [Google Scholar]
- Huez G., Marbaix G., Hubert E., Cleuter Y., Leclercq M., Chantrenne H., Devos R., Soreq H., Nudel U., Littauer U. Z. Readenylation of polyadenylate-free globin messenger RNA restores its stability in vivo. Eur J Biochem. 1975 Nov 15;59(2):589–592. doi: 10.1111/j.1432-1033.1975.tb02486.x. [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., Brawerman G. Characterization of the steady-state population of messenger RNA and its poly(adenylic acid) segment in mammalian cells. Biochemistry. 1974 Oct 22;13(22):4633–4637. doi: 10.1021/bi00719a026. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kindy M. S., Sonenshein G. E. Regulation of oncogene expression in cultured aortic smooth muscle cells. Post-transcriptional control of c-myc mRNA. J Biol Chem. 1986 Sep 25;261(27):12865–12868. [PubMed] [Google Scholar]
- Knight E., Jr, Anton E. D., Fahey D., Friedland B. K., Jonak G. J. Interferon regulates c-myc gene expression in Daudi cells at the post-transcriptional level. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1151–1154. doi: 10.1073/pnas.82.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska A., Brawerman G. Structural features in the 3'-terminal region of polyribosome-bound rabbit globin messenger RNAs. J Biol Chem. 1986 Jan 5;261(1):397–402. [PubMed] [Google Scholar]
- Krowczynska A., Yenofsky R., Brawerman G. Regulation of messenger RNA stability in mouse erythroleukemia cells. J Mol Biol. 1985 Jan 20;181(2):231–239. doi: 10.1016/0022-2836(85)90087-7. [DOI] [PubMed] [Google Scholar]
- Kwan S. W., Brawerman G. A particle associated with the polyadenylate segment in mammalian messenger RNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3247–3250. doi: 10.1073/pnas.69.11.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Hatton K. S., Skoultchi A. I., Schildkraut C. L. c-myc mRNA levels in the cell cycle change in mouse erythroleukemia cells following inducer treatment. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5323–5327. doi: 10.1073/pnas.82.16.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A., Pattengale P. K., Kuo A., Stewart T. A., Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986 May 23;45(4):485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. A., McCormack J. E., Buckler A., Sonenshein G. E. Transcriptional and posttranscriptional control of c-myc gene expression in WEHI 231 cells. Mol Cell Biol. 1986 Nov;6(11):4112–4116. doi: 10.1128/mcb.6.11.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan D. E., Osley M. A., Hereford L. M. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):614–621. doi: 10.1128/mcb.7.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrow R. E., Jacobson A. Increased rates of decay and reduced levels of accumulation of the major poly(A)-associated proteins of Dictyostelium during heat shock and development. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1858–1862. doi: 10.1073/pnas.84.7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford R. M., Wydro R. M., Nguyen H. T., Nadal-Ginard B. Cytoplasmic processing of myosin heavy chain messenger RNA: evidence provided by using a recombinant DNA plasmid. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5749–5753. doi: 10.1073/pnas.77.10.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J. F., Wake S. A. An analysis of the rate of metallothionein mRNA poly(A)-shortening using RNA blot hybridization. Nucleic Acids Res. 1985 Nov 25;13(22):7929–7943. doi: 10.1093/nar/13.22.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel C. G., Kwan S., Lingrel J. B. Size of the polyadenylic acid region of newly synthesized globin messenger ribonucleic acid. J Biol Chem. 1975 May 25;250(10):3725–3728. [PubMed] [Google Scholar]
- Miyamoto C., Chizzonite R., Crowl R., Rupprecht K., Kramer R., Schaber M., Kumar G., Poonian M., Ju G. Molecular cloning and regulated expression of the human c-myc gene in Escherichia coli and Saccharomyces cerevisiae: comparison of the protein products. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7232–7236. doi: 10.1073/pnas.82.21.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschel R., Khoury G., Reid L. M. Regulation of insulin mRNA abundance and adenylation: dependence on hormones and matrix substrata. Mol Cell Biol. 1986 Jan;6(1):337–341. doi: 10.1128/mcb.6.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H., Maxwell I. H., Pizer L. I. Herpesvirus infection alters the steady-state levels of cellular polyadenylated RNA in polyoma virus-transformed BHK cells. J Virol. 1982 Jun;42(3):1131–1134. doi: 10.1128/jvi.42.3.1131-1134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath P., Getzenberg R., Beebe D., Pallansch L., Zelenka P. c-myc mRNA is elevated as differentiating lens cells withdraw from the cell cycle. Exp Cell Res. 1987 Mar;169(1):215–222. doi: 10.1016/0014-4827(87)90239-4. [DOI] [PubMed] [Google Scholar]
- Nemer M., Dubroff L. M., Graham M. Properties of sea urchin embryo messenger RNA containing and lacking poly(A). Cell. 1975 Oct;6(2):171–178. doi: 10.1016/0092-8674(75)90007-0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel U., Soreq H., Littauer U. Z. Globin mRNA species containing poly(A) segments of different lengths. Their functional stability in Xenopus oocytes. Eur J Biochem. 1976 Apr 15;64(1):115–121. doi: 10.1111/j.1432-1033.1976.tb10279.x. [DOI] [PubMed] [Google Scholar]
- Paek I., Axel R. Glucocorticoids enhance stability of human growth hormone mRNA. Mol Cell Biol. 1987 Apr;7(4):1496–1507. doi: 10.1128/mcb.7.4.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik C. M., Wilkins C., Jacobson A. Translational control during early Dictyostelium development: possible involvement of poly(A) sequences. Cell. 1984 Apr;36(4):1017–1025. doi: 10.1016/0092-8674(84)90051-5. [DOI] [PubMed] [Google Scholar]
- Pellegrini S., Basilico C. Rat fibroblasts expressing high levels of human c-myc transcripts are anchorage-independent and tumorigenic. J Cell Physiol. 1986 Jan;126(1):107–114. doi: 10.1002/jcp.1041260115. [DOI] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Kobs G., Ross J. Substrate specificity of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987 Jul 5;262(19):9382–9388. [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts P. H., Forster A., Stinson M. A., Rabbitts T. H. Truncation of exon 1 from the c-myc gene results in prolonged c-myc mRNa stability. EMBO J. 1985 Dec 30;4(13B):3727–3733. doi: 10.1002/j.1460-2075.1985.tb04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts P. H., Watson J. V., Lamond A., Forster A., Stinson M. A., Evan G., Fischer W., Atherton E., Sheppard R., Rabbitts T. H. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. EMBO J. 1985 Aug;4(8):2009–2015. doi: 10.1002/j.1460-2075.1985.tb03885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmsdorf H. J., Schönthal A., Angel P., Litfin M., Rüther U., Herrlich P. Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res. 1987 Feb 25;15(4):1643–1659. doi: 10.1093/nar/15.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G., Evan G. I., Bishop J. M. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran W., Dean M., Levine R. A., Henkle C., Campisi J. Induction of c-fos and c-myc mRNA by epidermal growth factor or calcium ionophore is cAMP dependent. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8216–8220. doi: 10.1073/pnas.83.21.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Meneceur P., Tavitian A., Robert-Lezenes J. Presence of a c-myc transcript initiated in intron 1 in Friend erythroleukemia cells and in other murine cell types with no evidence of c-myc gene rearrangement. Mol Cell Biol. 1987 Feb;7(2):940–945. doi: 10.1128/mcb.7.2.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo L. L., Guild G. M. Poly(A) shortening of coregulated transcripts in Drosophila. Dev Biol. 1986 Jun;115(2):507–510. doi: 10.1016/0012-1606(86)90271-x. [DOI] [PubMed] [Google Scholar]
- Ross J., Kobs G., Brewer G., Peltz S. W. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987 Jul 5;262(19):9374–9381. [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Ross J., Peltz S. W., Kobs G., Brewer G. Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3' terminus. Mol Cell Biol. 1986 Dec;6(12):4362–4371. doi: 10.1128/mcb.6.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Pizarro A. Human beta and delta globin messenger RNAs turn over at different rates. J Mol Biol. 1983 Jul 5;167(3):607–617. doi: 10.1016/s0022-2836(83)80101-6. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W., Kornberg R. D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987 Sep;7(9):3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Hayday A. C., Wiman K., Hayward W. S., Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejersen T., Sümegi J., Ringertz N. R. Density-dependent arrest of DNA replication is accompanied by decreased levels of c-myc mRNA in myogenic but not in differentiation-defective myoblasts. J Cell Physiol. 1985 Dec;125(3):465–470. doi: 10.1002/jcp.1041250315. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Sheiness D., Darnell J. E. Polyadenylic acid segment in mRNA becomes shorter with age. Nat New Biol. 1973 Feb 28;241(113):265–268. doi: 10.1038/newbio241265a0. [DOI] [PubMed] [Google Scholar]
- Sheiness D., Puckett L., Darnell J. E. Possible relationship of poly(A) shortening to mRNA turnover. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1077–1081. doi: 10.1073/pnas.72.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H., Sagar A. D., Sehgal P. B. Translational activity and functional stability of human fibroblast beta 1 and beta 2 interferon mRNAs lacking 3'-terminal RNA sequences. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1741–1745. doi: 10.1073/pnas.78.3.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunitha I., Slobin L. I. An in vitro system derived from Friend erythroleukemia cells to study messenger RNA stability. Biochem Biophys Res Commun. 1987 Apr 29;144(2):560–568. doi: 10.1016/s0006-291x(87)80003-7. [DOI] [PubMed] [Google Scholar]
- Swartwout S. G., Preisler H., Guan W. D., Kinniburgh A. J. Relatively stable population of c-myc RNA that lacks long poly(A). Mol Cell Biol. 1987 Jun;7(6):2052–2058. doi: 10.1128/mcb.7.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. V., Gusse M., Evan G. I., Dathan N., Mechali M. Xenopus myc proto-oncogene during development: expression as a stable maternal mRNA uncoupled from cell division. EMBO J. 1986 Dec 20;5(13):3563–3570. doi: 10.1002/j.1460-2075.1986.tb04683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Watson D. K., Psallidopoulos M. C., Samuel K. P., Dalla-Favera R., Papas T. S. Nucleotide sequence analysis of human c-myc locus, chicken homologue, and myelocytomatosis virus MC29 transforming gene reveals a highly conserved gene product. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3642–3645. doi: 10.1073/pnas.80.12.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Welsh M., Nielsen D. A., MacKrell A. J., Steiner D. F. Control of insulin gene expression in pancreatic beta-cells and in an insulin-producing cell line, RIN-5F cells. II. Regulation of insulin mRNA stability. J Biol Chem. 1985 Nov 5;260(25):13590–13594. [PubMed] [Google Scholar]
- Westin E. H., Wong-Staal F., Gelmann E. P., Dalla-Favera R., Papas T. S., Lautenberger J. A., Eva A., Reddy E. P., Tronick S. R., Aaronson S. A. Expression of cellular homologues of retroviral onc genes in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2490–2494. doi: 10.1073/pnas.79.8.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. T., Wilson L. B., deRiel J. K., Villa-komaroff L., Efstratiadis A., Forget B. G., Weissman S. M. Insertion of synthetic copies of human globin genes into bacterial plasmids. Nucleic Acids Res. 1978 Feb;5(2):563–581. doi: 10.1093/nar/5.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. C., Sawicki S. G., White P. A., Darnell J. E., Jr A correlation between the rate of poly(A) shortening and half-life of messenger RNA in adenovirus transformed cells. J Mol Biol. 1978 Nov 25;126(1):23–36. doi: 10.1016/0022-2836(78)90277-2. [DOI] [PubMed] [Google Scholar]
- Zeevi M., Nevins J. R., Darnell J. E., Jr Newly formed mRNA lacking polyadenylic acid enters the cytoplasm and the polyribosomes but has a shorter half-life in the absence of polyadenylic acid. Mol Cell Biol. 1982 May;2(5):517–525. doi: 10.1128/mcb.2.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- Zullo J. N., Cochran B. H., Huang A. S., Stiles C. D. Platelet-derived growth factor and double-stranded ribonucleic acids stimulate expression of the same genes in 3T3 cells. Cell. 1985 Dec;43(3 Pt 2):793–800. doi: 10.1016/0092-8674(85)90252-1. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]