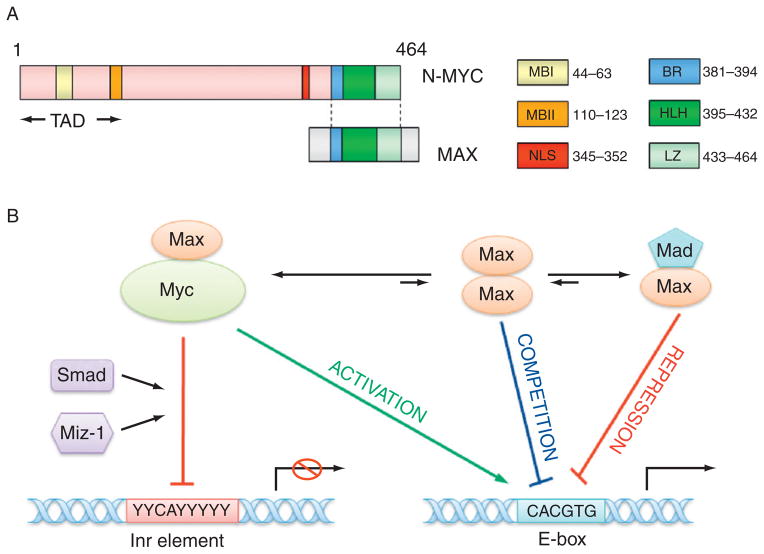
Figure 4. 3.
The structure of N-myc protein and transcription regulation by N-myc. (A) The structure of N-myc protein. The N-terminal transactivation domain (TAD) contains two conserved Myc box I and II (MBI and MBII), which are essential for DNA binding. The C-terminal domain (CTD) harbors basic region (BR), helix-loop-helix (HLH) motif, and leucine zipper (LZ) for dimerization with Max. There is a nuclear localization signal (NLS) before CTD. (B) The model for the transcription regulation by N-myc. Myc–Max heterodimer may bind to E-box element (CACGTG) to activate transcription, however, Myc–Max dimer can associate with other transcription factors such as Miz-1, Smad, and bind to Inr (initiator, weak consensus) element to repress transcription. Max can also form homodimers or heterodimers with Mad to compete or suppress Myc–Max binding to E-box.