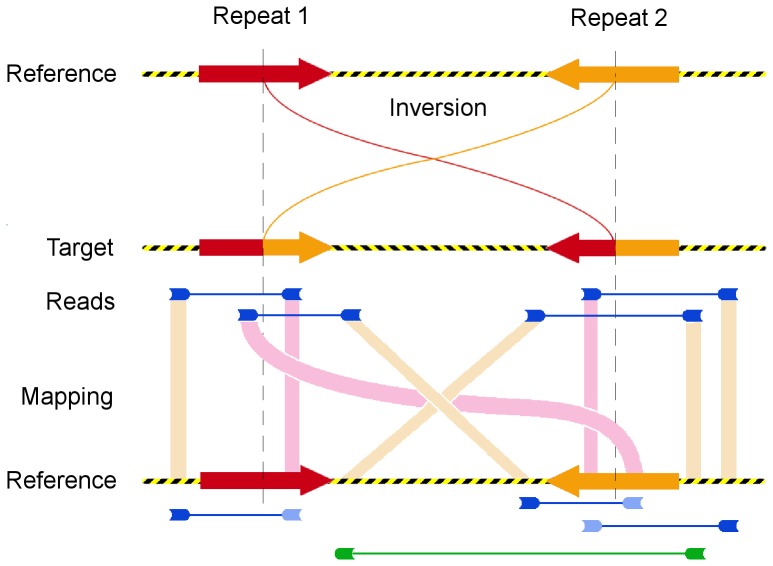
Figure 1. Inversion between the reference and the target genomes.
The breakpoints (dashed lines) are located inside inverted repeats (red or orange arrows). Four pairs of reads that span the breakpoints are depicted in blue, with their sequenced ends in opposite orientations. Yellow bands indicate the correct mappings in the reference genome of ends located in unique sequences. The reads sequenced from a repeat are erroneously mapped to the alternative copy (pink bands), because concordant alignments are favored by the aligner. The mapped reads at the bottom are displayed in dark blue if correctly mapped or in light blue otherwise. The only discordant pair of reads that report the inversion is shown in green.