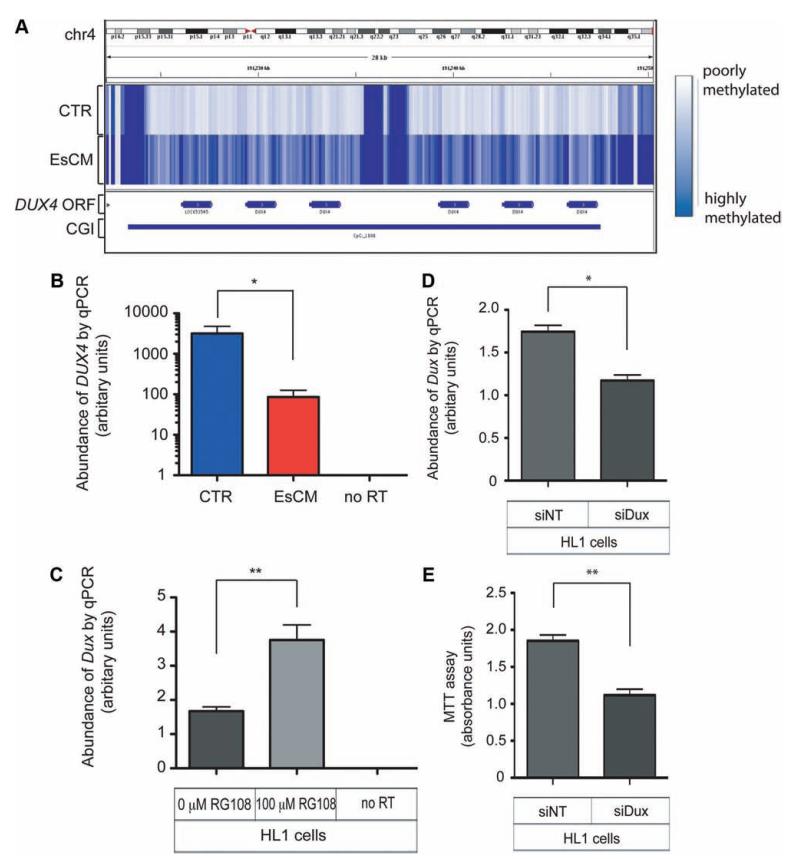
Figure 6.
Differential DNA methylation and expression of DUX4 in human hearts and Dux in the mouse HL1 cardiac cell line. A, The intronless DUX4 open reading frame is embedded within the subtelomeric array of D4Z4 repeat units. The CpG island (CGI) in this locus is hypermethylated in end-stage cardiomyopathy (EsCM) hearts but not in control (CTR). B, The abundance of DUX4 RNA transcripts was quantified with DNase-treated RNA from a panel of left ventricular tissue (8 control and 16 EsCM; Table I in the online-only Data Supplement) and normalized by geNorm that was generated with 2 housekeeping genes, RPLPO and TBP. Absent polymerase chain reaction (PCR) products in “no reverse transcriptase (RT) ” controls excluded the likelihood of genomic DNA amplification. *P<0.05. C, The mouse HL1 cardiac cell line was cultured with or without the specific inhibitor of DNA methyltransferase RG108 for 48 hours, and Dux RNA abundance was quantified with DNase-treated RNA and normalized to the housekeeping gene Gapdh (n=3; **P<0.01). D, HL1 cardiac cells were transfected for 48 hours with either control nontargeting siRNA (siNT) or siRNA targeting mouse Dux (siDux). Dux abundance was quantified as in C (n=3; *P<0.05). E, Methylthiazolyldiphenyl–tetrazolium bromide (MTT) assay was performed as described in Methods and represents survival of HL1 cells 48 hours after transfection with either siNT or siDux (n=3; **P<0.01). Statistical analyses for quantitative PCR (qPCR) and the MTT assay were by the Student t test, and all error bars represent the SEM.