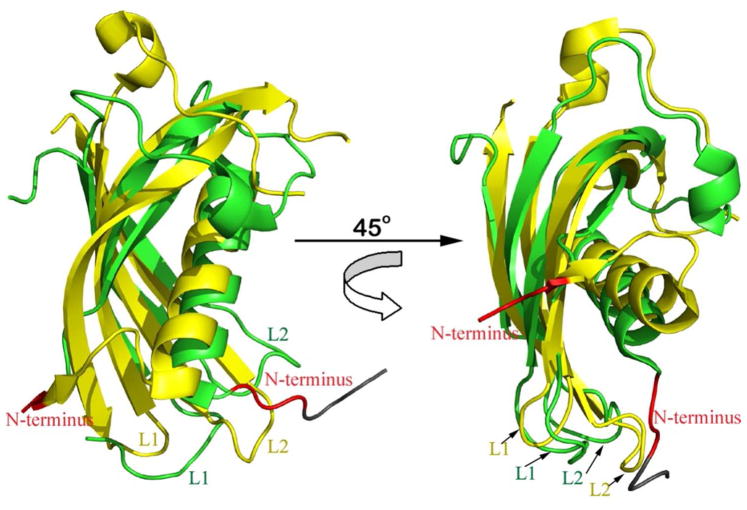
Figure 6.
Structural alignment of hCLD and the cysteine protease inhibitor Cystatin C. Backbone atoms of hCLD were superimposed on monomeric human Cystatin C in the unliganded state (PDB code: 3GAX). hCLD is shown in green, Cystatin C in yellow and, in both, the N-terminus is colored in red. Four additional residues at the N-terminus of hCLD as remnants of the cleavage by thrombin are colored in grey. The crystal structure of monomeric human cystatin C stabilized against aggregation was chosen for comparison since in all crystal structures of Cystatin C studied to date, the protein has been found to form 3D domain-swapped dimers, created through a conformational change of a beta-hairpin loop, L1 (68, 88–90). 3D domain swapping free structure defines the conformation of loop L1, which is essential for the inhibition of papain-like cysteine proteases.