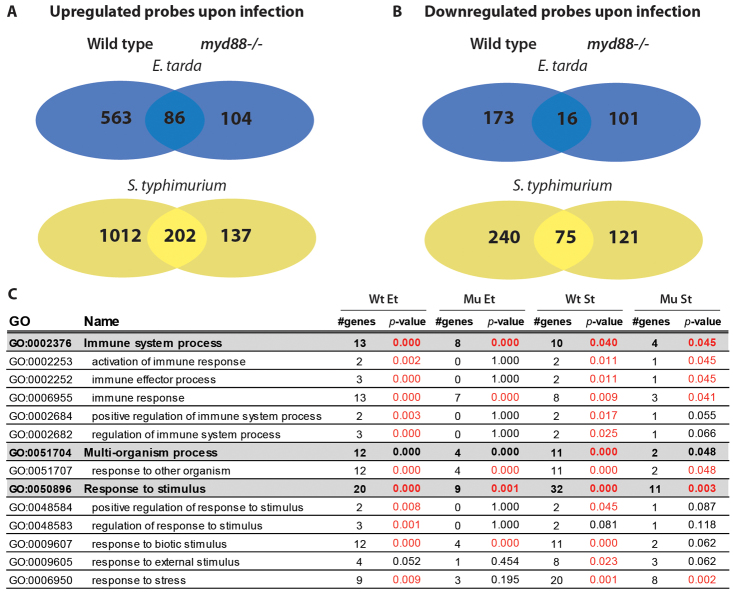
Fig. 2.
Transcriptome analysis of myd88−/− and wild-type embryos following infection with E. tarda and S. typhimurium. (A,B) Venn diagrams showing the overlap and differences between mutant and wild-type embryos in numbers of probes upregulated (A) or downregulated (B) by E. tarda FL6-60 (Et) or S. typhimurium SL1027 (St) infection. Embryos were infected with ∼150 cfu of either pathogen into the caudal vein at 28 hpf and snap-frozen individually at 8 hpi. Triplicate samples for each infection condition were compared groupwise with samples from control embryos (injected with PBS) using a common reference microarray design. Significance cut-offs for the ratios of infected versus control groups were set at twofold with P<10−5. (C) GO analysis. The table displays the enrichment of immune-related GO terms following infection of myd88−/− (Mu) and wild-type (Wt) embryos with E. tarda (Et) or S. typhimurium (St). GO-term enrichment in the gene lists upregulated by infection was determined by master-target statistical testing using eGOn (Beisvag et al., 2006) with the Danio rerio UniGene build number 124 as background. The numbers of genes associated with each GO term and the P-values for enrichment are indicated. We note that GO analysis is limited by the number of Unigene clusters annotated for zebrafish and therefore not all upregulated probes could be included in the analysis.