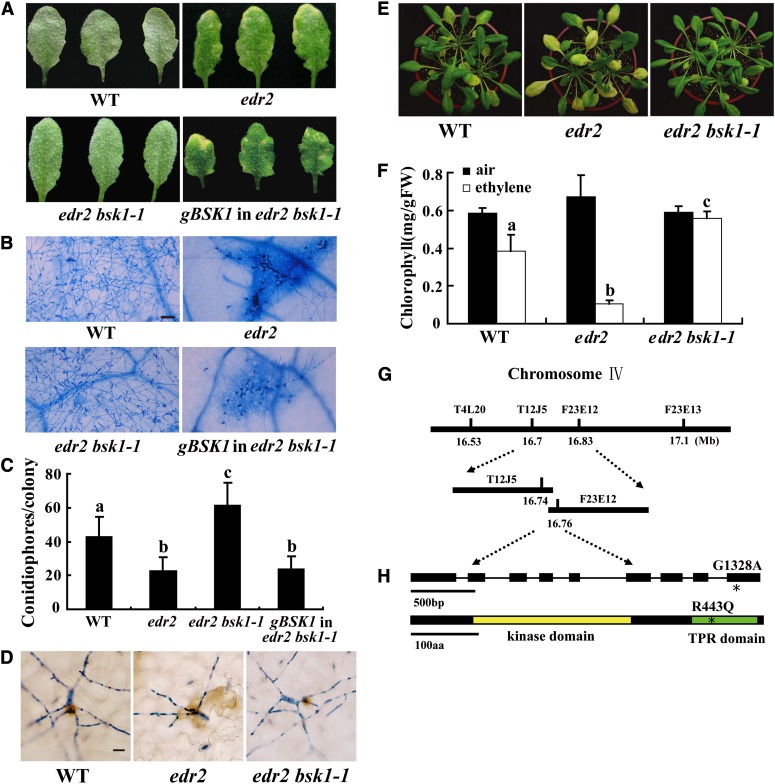
Figure 1.
Suppression of the edr2-Mediated Phenotypes by bsk1-1.
(A) Four-week-old Arabidopsis plants were infected with G. cichoracearum. The plants were photographed at 8 DAI. The wild-type plants were susceptible, and a large number of spores were produced on the leaves. By contrast, the edr2 mutant was resistant, displaying massive necrotic cell death and very few spores. The edr2 bsk1-1 plants displayed a wild-type-like susceptible phenotype, and no visible necrotic lesions were present on the leaves. A genomic clone containing the BSK1 gene complemented the bsk1-1 mutation. WT, the wild type.
(B) Fungal growth and cell death on 4-week-old plants at 8 DAI with G. cichoracearum. Leaves were stained with trypan blue to show fungal structures and dead cells. A large number of spores were produced in the wild type and edr2 bsk1-1, while extensive mesophyll cell death occurred in edr2. Bar = 50 μm
(C) Quantification of fungal growth in plants at 5 DAI by counting the number of conidiophores per colony. Results were from one experiment, and the bars represent mean and sd (n > 30). Statistically significant differences are indicated by lowercase letters (P < 0.05; one-way analysis of variance [ANOVA]). The experiments were repeated three times with similar results.
(D) Infected leaves were stained with 3,3′-diamino benzidine-HCl and trypan blue sequentially at 2 DAI to visualize H2O2 (brown staining) and fungal structures (blue staining). Bar = 20 μm
(E) and (F) The bsk1-1 mutation suppressed enhanced ethylene-induced senescence in edr2. Four-week-old plants were exposed to 100 μL L−1 ethylene for 3 d, and increased chlorosis was observed in edr2 mutants after ethylene treatment. The edr2 bsk1-1 plants showed a wild-type-like phenotype (E). The chlorophyll content in leaves from the ethylene treated plants (F). Bars represent sd of values obtained from five plants. Lowercase letters represent significant difference from the wild type (P < 0.01, one-way ANOVA). Three independent experiments were performed with similar results. FW, fresh weight.
(G) The bsk1-1 mutation was identified by standard map-based cloning. Markers and BAC clones are indicated.
(H) Structure of the BSK1 gene. The asterisk indicates the bsk1-1 mutation. Exons are indicated by black boxes and introns by black lines. A nucleotide change (G1328A) was identified in the BSK1 gene and leads to an amino acid substitution (R443Q) in the BSK1 protein. The BSK1 protein contains a kinase domain and a TPR domain. The bsk1-1 point mutation (asterisk) is in the TPR domain. aa, amino acids.