Figure 4.
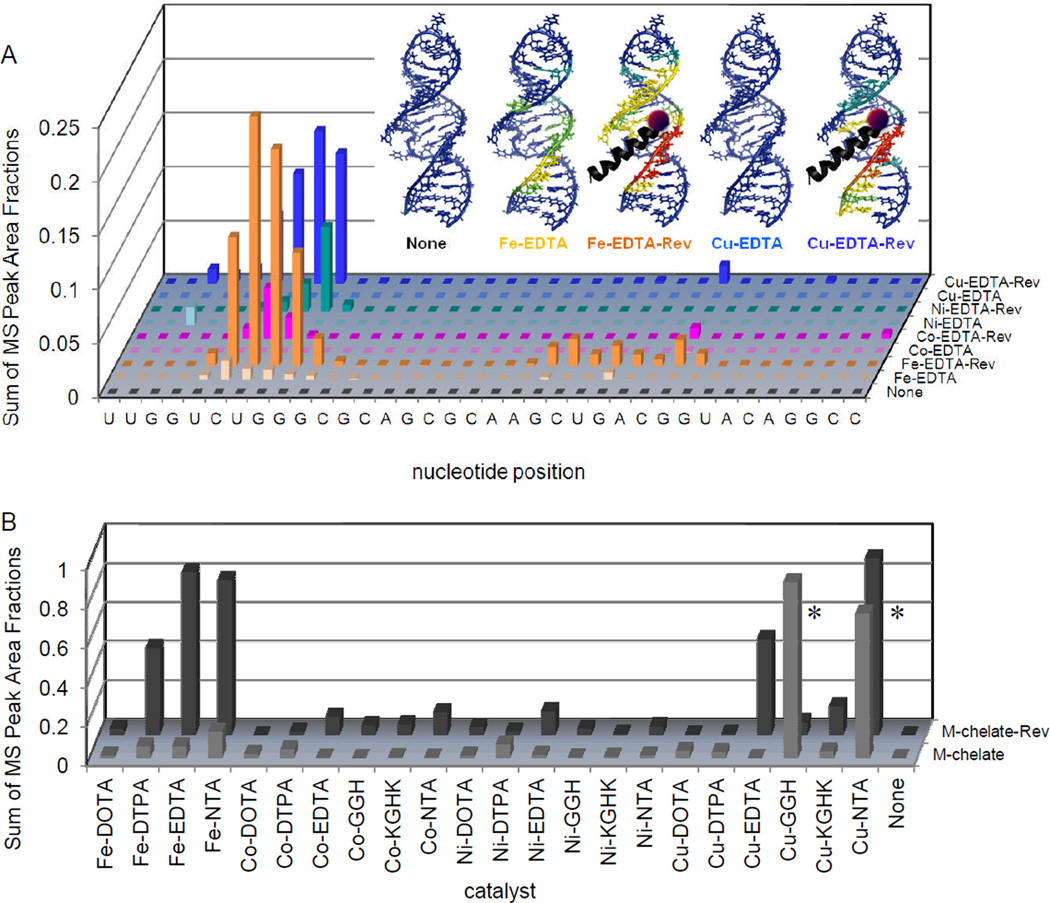
Targeted cleavage of RRE RNA required attachment of catalysts to the Rev peptide in order to achieve efficient cleavage, as seen by differences in reactivity between the M-chelate-Rev catalysts and M-chelates lacking Rev. After 1 h incubation of each catalyst (10 µM) and co-reactants (1 mM ascorbate/H2O2) with the Fl-RRE RNA (10 µM), the apparent abundances of individual RNA cleavage fragments were quantified. (A) The data correspond to the abundances (MS peak area fractions) of oxidation products (cleavage fragments containing nascent terminal 3’-PO4, 3’-PG, or 5’-PO4 overhangs), at positions adjacent to each illustrated site of H-abstraction, after 1h reaction. Inset: The same data were mapped onto the 3D solution structure of the RRE RNA,10 for several catalysts and the control catalysts lacking the Rev targeting domain (red corresponds to highest reactivity; blue corresponds to reactivity below background). (B) Comparison of the reactivity of each M-chelate-Rev catalyst vs each M-chelate lacking Rev, in the presence of co-reactants, after 1h reaction. Plots similar to that shown in (A) are shown for all catalysts in Figures SM15 to SM17 of the Supporting Information. * The relatively high activity of the Cu-GGH and Cu-NTA chelates lacking Rev was the result of the combination of a low-affinity Cu-binding site within the RRE RNA and the high concentrations of catalyst and RNA required for MALDI-TOF MS detection.