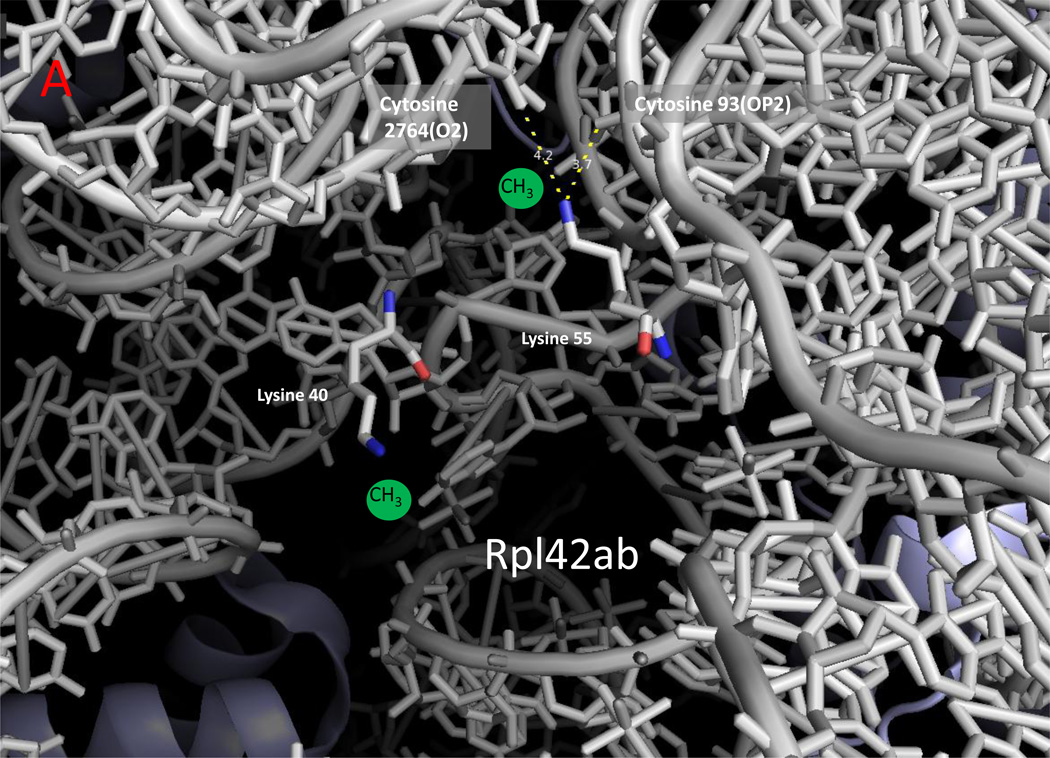
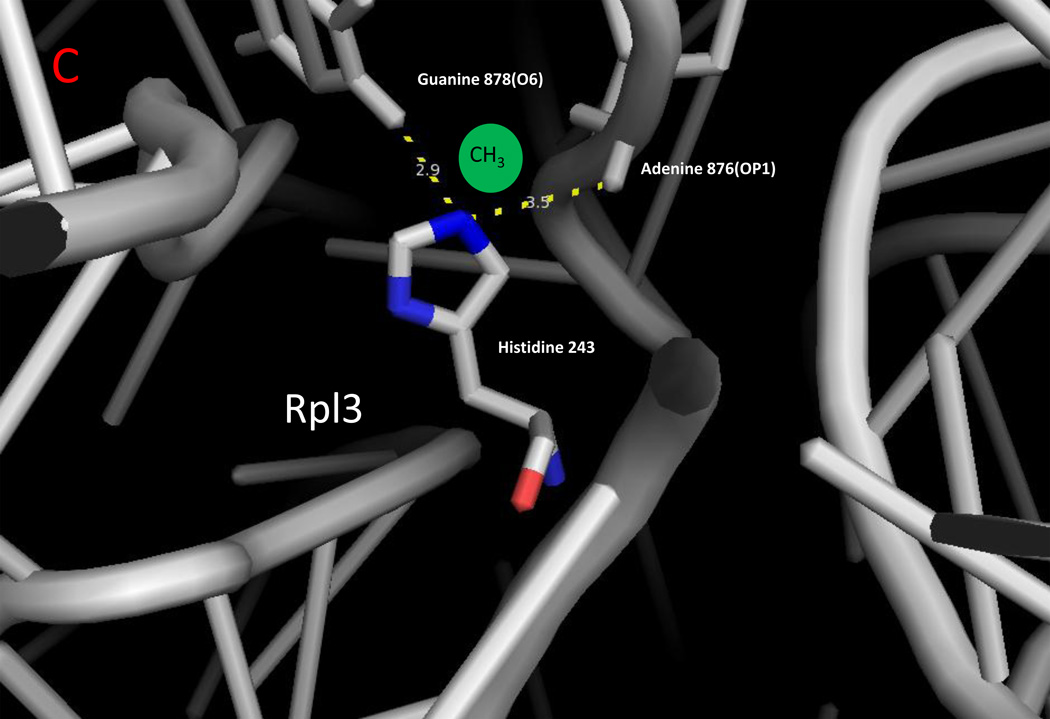
Figure 2.
Zoomed in view of methylated lysine, arginine, and histidine residues with ribosomal RNA in yeast cytoplasmic ribosomes. The monomethylated residues of Rpl42ab ((a); Lys-39; Lys-54), Rps3 ((b); Arg-145), and Rpl3 ((c); His-242) are shown with nitrogen atoms in blue and oxygen atoms in red [8,14,25]. Ribosomal 25S (Panels A and C) and 18S (Panel B) RNA are shown in gray. Methyl groups were not modeled into these structures; possible locations are suggested by the green spheres. Examples of close contact distances (less than 5 Å) are shown between the methylated atom in the protein and the RNA to emphasize the apposition of the methylated residue to the RNA, although these interactions may differ in a refined structure that includes the methyl groups. The illustration was made using PyMOL and the PDB structures 3U5F, 3U5G, 3U5H, and 3U5I [45].