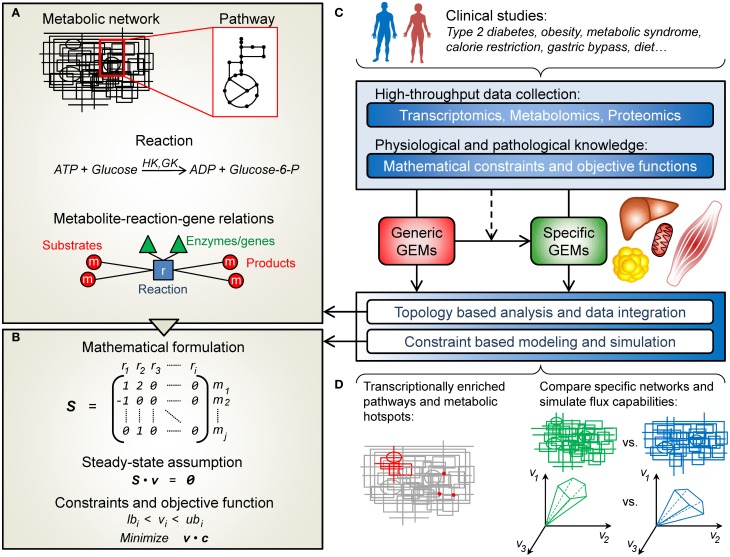
Figure 1.
An overview of human genome-scale metabolic models (GEMs) and their applications in the field of obesity and diabetes. (A) A metabolic network is in simple terms a list of the chemical reactions taking place in a cell. These reactions can be grouped into pathways and associated with a particular cellular compartment (e.g., mitochondria). Metabolites can be passed between compartments through transport reactions. Each reaction can be associated to its corresponding enzyme-coding genes, and together all the reactions provide a network structure connecting metabolites, reactions and genes. (B) The metabolic network can be represented mathematically by the stoichiometric matrix, S, containing the stoichiometric coefficients of the metabolites (rows) taking part in each reaction (columns). Under the constraint based modeling framework it is assumed that the metabolite concentrations are unchanged (Sv = 0). Further on, additional constraints can be put on the flux vector, v, to find capable and probable flux distributions. Alternatively, flux balance analysis (FBA) can be used to find a flux vector that optimizes an objective function (e.g., maximize ATP production). (C) GEMs have been used to study obesity- and diabetes-related conditions. Clinical data can be used to construct context specific GEMs from generic ones. This type of data can also be integrated and analyzed, in combination or separately, with the GEMs, in a topological or simulation based manner. (D) This enables e.g., the identification of transcriptionally affected reactions and pathways as well as metabolic hotspots, or the comparison of simulation results in terms of network capabilities.