Figure 2.
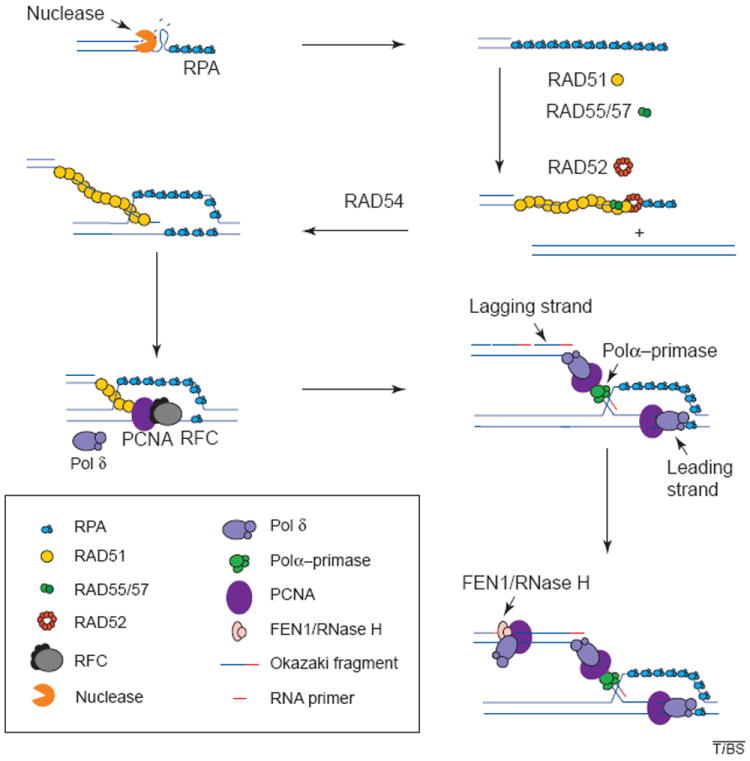
Recombination and associated DNA synthesis. The resection of the ends of the double strand creates a 3′ single-stranded DNA molecule. The binding of RPA to the single strand protects the DNA from degradation and disrupts any secondary structure. RAD51 is then loaded onto the DNA strand with the aid of RAD52, RAD55 and RAD57, and in the presence of RAD54, the RAD51 filament carries out strand exchange. The primer generated is recognized by the polymerase (Pol) δ accessory factor RFC, which loads PCNA onto the primer end generating a complex that is recognized by Pol δ or ε, which will carry out synthesis of the leading strand. Lagging-strand synthesis initiates when RNA primers are synthesized on the leading strand by the primase activity of the Pol-α–primase complex and are immediately extended by the Pol α into short DNAs. The Pol δ (or ε) system is then assembled on these DNA primers, replacing Pol α. The primers are then extended to yield Okazaki fragments. These accumulate on the lagging strand until they mature by the combined action of FEN1 and RNase H nucleases, which remove the RNA moiety of the fragment, leaving a gap that is then filled in by DNA Pol. DNA ligase seals the resulting nick yielding an intact double-stranded DNA molecule.
