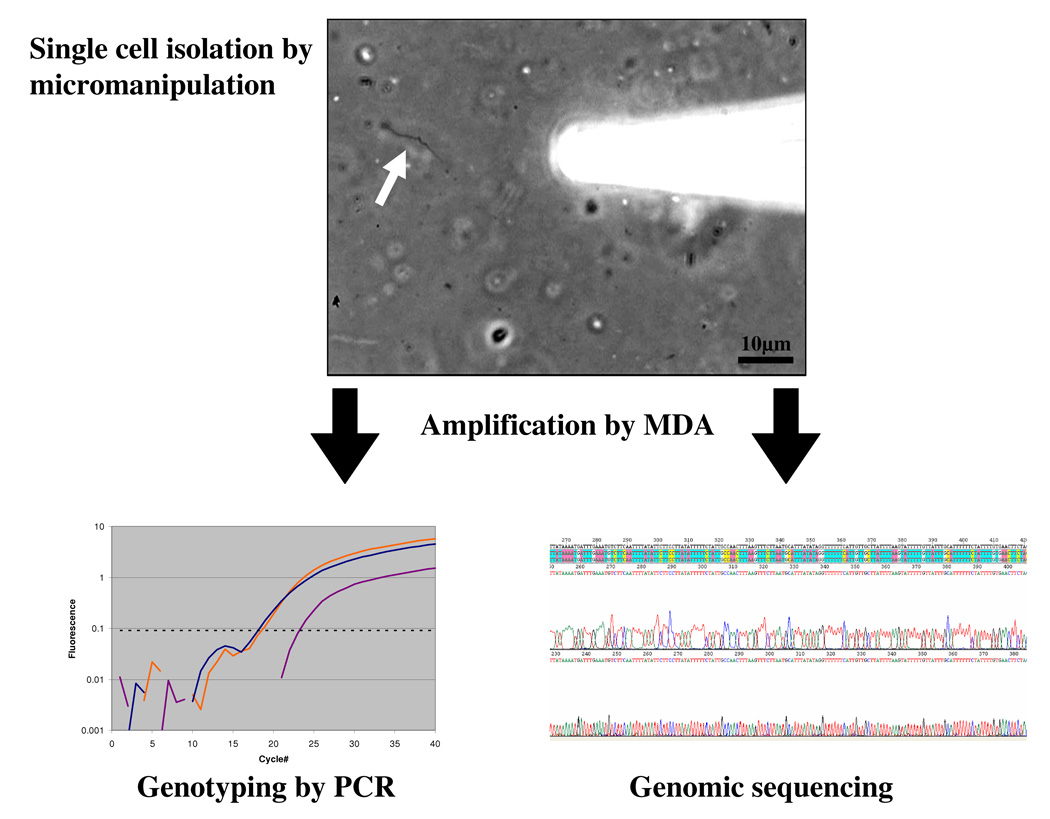
Figure 1.
Single cell MDA provides sufficient DNA to carry out PCR analysis of many loci. A single Borrelia cell (arrow) was isolated directly from tick midgut tissue by micromanipulation. The individual cells were amplified in 5μl MDA reactions yielding approximately 4.5μg DNA product which was used as template in PCR reactions with primers for loci previously employed for MLST analysis of cultured Borrelia isolates [49]. The PCR products were sequenced and confirmed the published genotypes of outer surface proteins (ospA, ospB, osp C). PCR using total DNA extracted from the tissue, an alternative approach, makes it impossible to determine if genotypes of different loci were genetically linked within individual cells, as the data obtained would be from all of the bacterial cells present. While culturing isolates is an alternative method that does provide linkage information, the organisms for which adequate culture conditions are available are very limited and there is often strong selection for fast growing strains biasing the results. Moreover, virulence factors associated with plasmids can be rapidly lost in culture [50].