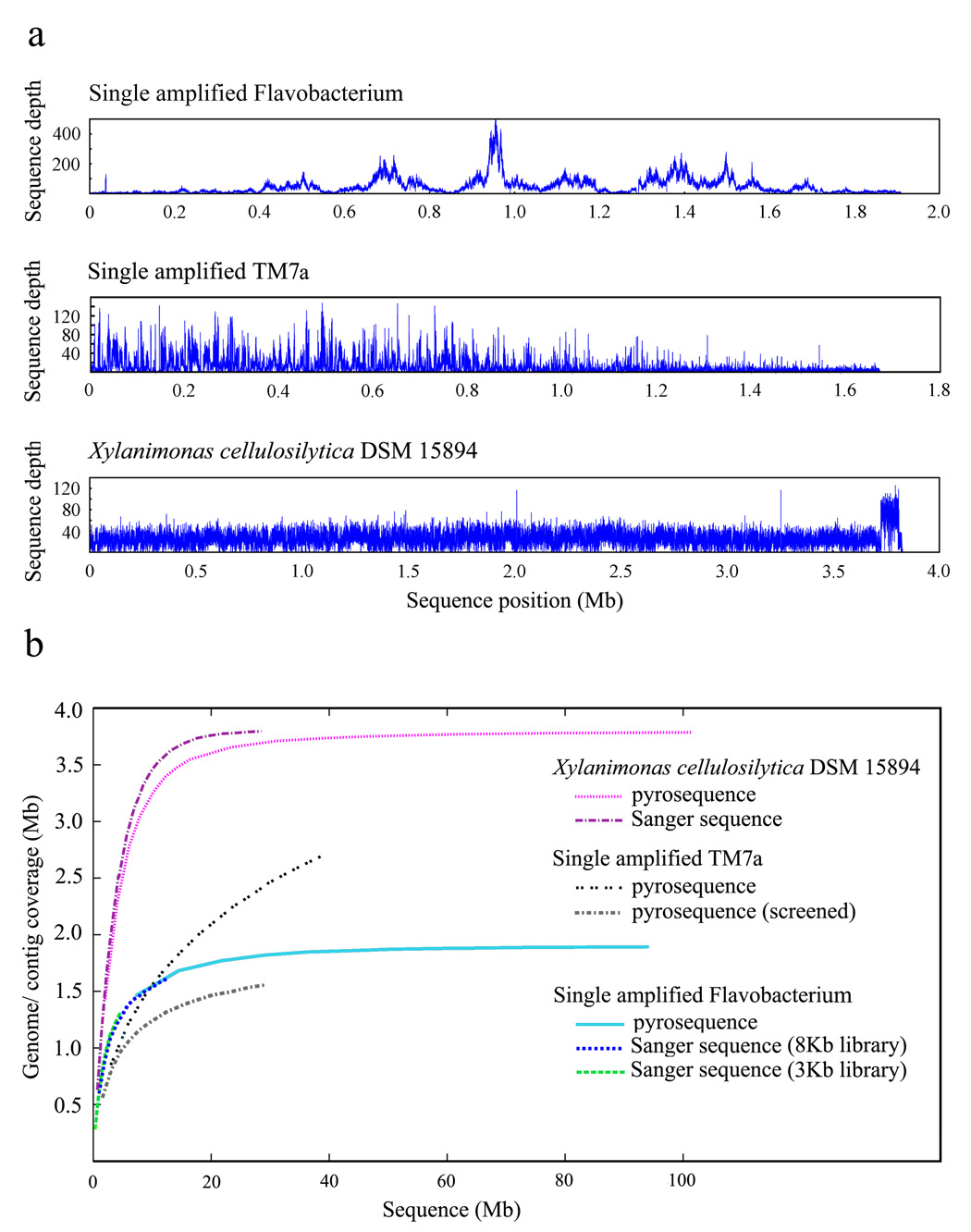
Figure 2.
Characterization of single cell MDA DNA sequence. (a) Pyrosequence depth distribution for a single amplified Flavobacterium (top panel) (TW, RS unpublished) and the single amplified TM7a (middle panel) [20] demonstrate MDA-originated high regional fluctuations in sequence coverage, while the sequence depth for the unamplified sample Xylanimonas cellulosilytica DSM 15894 exhibits a low degree of variation (bottom panel). Sequence positions were determined via arranging contigs by length. TM7a pyrosequence was screened to exclude reads contributing to contigs <300 bp and human reads. The mean sequence depth is 85.7 (± 73.7) for the flavobacterial SAG, 14.4 (± 16.2) for the TM7a SAG, and 31.9 (± 13.9) for X. cellulosilytica. A certain degree of sequence depth fluctuation may be attributed to repeats and mis-assemblies. (b) Genome coverage as function of the genome sequencing effort. Additional shotgun sequencing would not be effective in recovering the under-amplified genomic regions in the single amplified Flavobacterium (estimated genome size of ~2.3Mb) or TM7a (no genome estimate available). The X. cellulosilytica genome is estimated to be ~3.8Mb.